Biography
Interests
Samane Homaei, Farzane Farokhi* & Alam Ara Gholamis
Department of Biology, Faculty of Basic Sciences, Islamic Azad University, Sari Branch, Sari, Iran
*Correspondence to: Dr. Farzane Farokhi, Department of Biology, Faculty of Basic Sciences, Islamic Azad University, Sari Branch, Sari, Iran.
Copyright © 2019 Dr. Farzane Farokhi, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The term probiotic comes from Greek origin: pro and bios meaning “prolife”, which have used with different
meanings over the years [1]. It was used since 1970s to introduce the supplements fed with useful microbes for human
and animals [2]. The first study of probiotic bacteria from the aquaculture environment was reported in 1980s [3].
The use of probiotics for aquatic animals extremely demanded and must be harmless for aquaculture practices [4].
According to the reports of WHO, production of world aquaculture is noticeably growing and newly released FAO aquaculture statistics has recorded of 106 million tons in live production in the year 2015. Within 15 recent years, the aquaculture portion to the world production of aquatic animals (both captured and farmed) has increased from 25.7% in 2000 to 45.3% in 2015. Among all farmed aquatic animals for human feeding, aquaculture provided average 10.42kg of human eating food fish in 2015, which has been raising up to 0.28kg, from 10.14kg in 2014 and mainly by farming the finfish (trout) all around the world [5]. Fish diseases are major problem to be concerned and need urgent microbial control strategies in aquaculture because they have a great effect on aquaculture production, trading and occurring the antibiotic resistance [2].
It is predicted that global population will reach to 9 billion by the year 2050 and an important fact is that fish can play a key role in nourishing the world’s growing population with middle income. Already, fish contains 16 percent of all animal protein consuming in the world, then urgent attention is needed to overcome the aquaculture diseases. Amazingly probiotic bacteria can be active not only in intestinal tract but also in other parts like gills or skin of the host.
Adding probiotic bacteria to fishes' diet as supplements have many benefits including inhibition the growth of pathogenic microorganism's, increasing immune responses, increasing growth factors and enzymatic contribution to digestion [6].
Therefore, this study aimed to identifying and examining the inhibitory activity of Lactobacillus spp isolated from the finfish (trout), and evaluating the antagonistic activity against fresh water fish pathogenic bacteria Aeromonas Hydrophila.
Materials and Methods
40 healthy trout fishes, (ten from each pool) were obtained from 4 private fish pools in Mazandaran province,
Iran and transported to the laboratory in well aerated polythene bags. After euthanizing, their weight and
length were recorded carefully before dissection and the number of accidental organisms was reduced by
washing the fish skin with 70% ethanol. Then the ventral surface was opened with sterile scissors and
intestine cut longitudinal. After that, 1g of the intestinal tract content was removed under aseptic condition
and moved into previously weighed flasks containing storage medium [7].
Contents of Fishes’ intestines were homogenized in a storage medium using a vortex mixer and 1ml was
transferred to condensed neutralized bacteriological peptone (NBP, Oxoid L34, Hampshire, England)
Cysteine HCl 0.5g/L, NaCl 8g/L, pH adjusted to 6.7 [8].
Subsequently serial dilutions were spread on plates of selective culture media such as MRS agar (MRS, Merck, Darmstadt, Germany) with 1.5% agar (M641, HiMedia, Mumbai, India) and pH adjusted to 4.2 (MRS 4.2) and incubated anaerobically at 37ºC for 24-72 hour. Anaerobic incubation of the media was made in an anaerobic Gas-Pack system (LE002, HiMedia, Mumbai, India) with a mixture of 80% N2, 10% H2 and 10% CO2. The bacterial colonies were observed and sub cultured for further characterization and identification. Identified strains of lactobacilli were kept in MRS broth with 15% (v/v) glycerol at -20°C.
Strains were randomly selected for identification based on phonotypical characteristics. Cell morphology
and motility of all isolates were observed using a phase contrast microscope (CH3-BH-PC, Olympus,
Japan). The morphology of colonies as color, size and margin were recorded and subjected to Gram staining,
motility and biochemistry tests (eg., oxidase and catalase activity) followed by Barrow and Feltman (1993)
[9] key identification. Preliminary identification and grouping was based on the cell morphology and
phenotypic properties such as CO2 production from glucose, hydrolysis of arginine, growth at different
temperatures (10, 15 and 45ºC). All isolates were cultured on Mac Conky agar and Acetate agar (PH=5.4) to identify from other Gram negative and enteric bacteria.
Molecular Identification
Chromosomal DNA of bacteria was extracted directly from strain isolates [10]. Genomic DNA was prepared
by using the following procedure (Cardinal et al.,1997). Ten ml overnight cultures were prepared in MRS
broths. Cells were harvested in a micro centrifuge for 2min at 1300rpm. Afterward, they were suspended
in 200μl 1xTE buffer (pH8) containing 25% sucrose and 30mg/ml lysozyme. The cell suspensions were
then incubated for 1h at 37ºC. After the incubation, 370μl, 1x TE (pH 8) containing proteinase K (1mg/ml) and 30μl, 10% SDS were added. The samples were then incubated for 1h at 37ºC. Cells were lysed by
adding 100μl of 5M NaCl and 80μl CTAB/NaCl solution (10% cetytrimethylammonium bromide, 0.7 M
NaCl), respectively. Lysed samples were incubated for 10min at 65ºC. Chloroform extraction performed
twice (chloroform/ isoamyl alcohol: 24/1). First, one equal volume of chloroform/isoamyl alcohol added
and the samples were centrifuged for 10min at 13000rpm. The aqueous phase transferred into a new
eppendorf tube and the genomic DNA was precipitated by the adding isopropanol (one equal volume). Then
precipitated DNA was transferred into a fresh eppendorf tube which contained 500μl 70% ethanol, and
washed. Once DNA precipitate was not visible, isopropanol containing samples were centrifuged for 10min
at 13000rpm to pellet genomic DNA. After washing, DNA was pelleted by centrifugation for 5min at
13000rpm. Ethanol was removed and the pellets were dried for 10 min at 37ºC. Dried pellets were dissolved
in 100μl 1xTE. After incubation for 1h at 37ºC, the sample volume was adjusted to 100μl with 1xTE. DNA
dissolved by alternating cold-heat shock (10min at 80ºC and 20min at -20ºC, twice). Dissolved genomic
DNA samples stored at -20ºC [11]. A 1μl volume of this prepared bacterial DNA added to 99μl of master
mix for amplification of the 380-bp 16S rRNA target [12].
Well-isolated colonies on MRS agar plates were used as a template for the PCR amplification of 16S rRNA gene.
Briefly, the well-characterized primers, RW01(nt;1170-1189)5`-AACTGGAGGAAGGTGGGGAT-
3`and DG74(nt;1522-1540)5`- AGGAGGTGATCCAACCGCA-3` were used to amplify a 380-bp
(bp) fragment that resides within a conserved region of the bacterial 16S rRNA gene which is flanked by
variable regions V8 and V9. The master mix consisted of 10mmol/L Tris-HCl, pH 8.3, 50mmol/L MgCl2,
200μmol/L of each dATP, dCTP, dUTP, dGTP, 1 unit (U) of UNG (Applied Biosystems, Foster City,
CA), 25μmol/l of each primer and 2.5U Taq polymerase (Promega, Madison, WI). Ten μl of the prepared
specimens added to 90μl of master mix. Amplification of DNA performed in a PCR System: T100TM
Thermal Cycler (Bio-Rad, USA) programed for 5min at 95ºC (initial denaturation) and 35 cycles of 45s at
94ºC (denaturation), 1m at 63ºC (annealing), 1min at 72ºC (extension) and 5min at 72ºC (final extension).
After 35 amplification cycles, 20μl of reaction product analyzed by agarose gel electrophoresis. The ethidiumbromide stained gel was evaluated for the presence of the 380bp DNA fragment compared to a 100-bp ladder molecular weight standard (Vilber-Lourmat, France) [13].
The 380-bp amplicons from isolates of bacteria were fully sequenced by using the ABI 377 gen sequencer
(Perkin-Elmer). PCR amplification efficiency of each sample was assessed by agarose gel electrophoresis
before sequencing. Once adequate amplification was documented, the sequence subjected to homology
search using BLAST- querying the GenBank databasehttp://www.ncbi.nlm.nih.gov/blast"> http://www.ncbi.nlm.nih.gov/blast (last accessed
March, 2011) of the National Center for Biotechnology Information (NCBI).
The well diffusion agar method used for the assay of antagonistic effect of Lactobacillus isolates and it′s
growth inhibitory activity against the fish pathogen Aeromonas hydrophila. Colonies of Aeromonas hydrophila
which obtained from 18h old MRS broth culture mixed with MRS agar (1.2% agar) and poured on sterile
petri dishes and incubated at 37ºC for 48 hours. Plate surface punched by Cork borer to make wells (9mm
diameter) on agar plate, to perform well diffusion assay. Then 100μl fluid contained filtered culture of
Lactobacillus isolate was introduced in a well. The plates incubated at 37°C for 24 to 48hrs and growth
inhibition zones (GIZ) recorded (Jayanth et al., 2001).
SPSS software version 16 used to analyze the data. One-way ANOVA test used and P<0.05 considered as
a significant difference.
Results
In order to identify LAB (lactic acid bacteria), phenotypic methods performed which included morphological
exams and physiological, biochemical and molecular tests.
Isolates were analyzed under the light microscope. At this step, cell shape (eg., cocci, ovoid, rod) and
arrangements (eg., diploid form, chain form, tetrad form) were examined after staining. From cultured forty
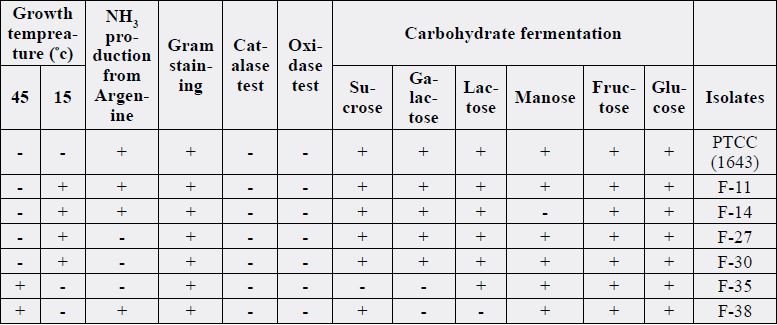
trout samples, finally twenty-five Gram positive cocci and bacilli form isolates identified.
From all twenty-five isolates, six isolates identified as Lactobacillus according to their motility, nitrate and
nitrite resuscitation, urease, growth in Mac Conky medium, difficulty in growth at acetate agar medium
(PH=4.5). Four isolates were enterococcus and five were corynebacterium. Six acid lactic bacteria and four non acid Isolates were analyzed under the light microscope. At this step, cell shape (eg., cocci, ovoid, rod)
and arrangements (eg., diploid form, chain form, tetrad form) were examined after staining. From cultured
forty trout samples, finally twenty-five Gram positive cocci and bacilli form isolates identified.
An approximately 20 bp region of the 16S rRNA gene was PCR-amplified from nine of the strains isolated
from fishes. The degree of sequence identity between the Lactobacillus species ranged from 88% to 99% over
the 380 bp (V8 and V9) used in the analysis.
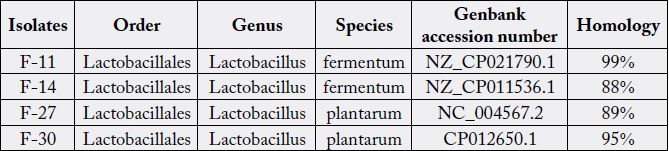
Sequences of unknown strains were individually aligned with those of the type strains. These alignments were edited to uniform length and a sequence identity matrix calculated for each of the unknown strains. Alignments of 16S rRNA gene sequences determined from the type strains divided the members into three groups including Lact. acidophilus, Lact. Plantarum and Lact. Fermentum (Table 2) (Figures 1 to 6).
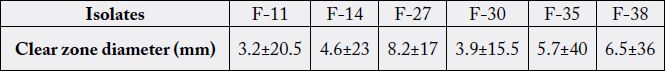
Six bacterial isolates were tested for their antibiosis against the fish pathogen Aeromonas hydrophila with gel
diffusion method. Results of antagonistic effect of six isolates are shown in Table 3. All six isolates expressed
antagonistic effect against the pathogen Aeromonas hydrophila. The best effect belonged to isolate F-35 (Lac.
acidophilus) while the isolate F-30(Lac. plantarum) showed the lowest antagonistic effect.
Discussion
Elie Metchnikoff (1908) was the first scientist who used the lactic acid bacteria (LAB) for human, (Vila et
al., 2010). It is over than 100 years that human have recognized the benefits of probiotic bacteria. Initially
Probiotics were used in diet to affect the growth rate and health situation of the host and increase the
resistance to diseases. Benefits of probiotic have been well demonstrated in human, poultry, pig and ruminant
nutrition, but the effect of such probiotics in aquatic animals is almost a new approach [6]. Bacterial infections
are an important cause of mortality in aquaculture farms [14]. Therapeutic strategies and prophylaxis
based on oral administration of chemical antimicrobial drugs and use of disinfectants in aquaculture farms
are the conventional methods [15]. The global production of farmed food fish was 59.9 million tons in
2010 [16]. By increased demand for aquaculture products, this section has changed and production has
increased to maximum range by adding commercial diets, antibiotics and other additives. Unfortunately,
all of these processes enforced the stressful conditions and consequently disease and economical loses [17].
For the first time, Aeromonas hydrophila was known in 1943. Aeromonas species as Gram-negative, nonspore-
forming, rod-shaped, facultative anaerobic bacteria with a widespread distribution, are found in water,
water habitants, domestic animals and foods [18]. A. hydrophila was identified as one of four Aeromonas
species. Also A.hydrophila as an emerging aquatic pathogen, has an important effect on environment. The
microorganism is a potential food- borne pathogen, especially hybridized strains which are responsible for
clinically ill cases [19,20]. Also A. hydrophila as a psychrotrophic microorganism is able to grow in foods
during refrigeration. Aeromonass species are able to excreting a number of extracellular toxins and enzymes
and the primary toxin are haemolysins, and the most significant of them is aerolysin, which is excreted by
many strains of A. hydrophila and A. sobria [21].
Aeromonas hydrophila can causes important infections in fish and generally associates with small lesions on fish body surface, ulcerative infections and hemorrhagic septicemia. These diseases are common in the world and produce considerable economic losses in aquaculture farming [22].
The disease is associated with spectrums such as gastroenteritis, septicemia, traumatic and aquatic wound infections, and infections after medical leech therapy [23]. Due to the Multiple resistance of the bacterium to several antimicrobial agents [24] an approach for inhibition of A. hydrophila growth, reported by Lewus, Kaiser, and Montville in 1991 [25]. The approach aimed to use bacteriocin bacteria that were isolated from meat cuts retail in order to produce lactic acid [26]. In another study, Santos et al, demonstrated that Lactococcus lactis sub sp, Lactis strain 388, inhibited the growth of A. hydrophila [27]. These researches imply the antagonism between A. hydrophila and lactic acid bacteria.
In recent years, "Probiotics" are used as useful bacteria for reducing bacterial/fungal infections in aquacultures. The potential advantages of this approach is that the intestinal microbial balance is saved and consequently it causes beneficial effects in host [16]. Lactobacillus with ability to adhere to the cells, exclude or reduce pathogenic adherence, produce lactic acid, persistence and multiplying, is considered a safe and non pathogen microorganism.
In present study Lactobacillus isolated from intestinal tract of trout fishes, were morphologically and biochemically characterized and identified with molecular method.
For molecular method, the 16S rRNA PCR assay was used, which can detect 10 to 50 colony forming units/ ml. Briefly, the well-characterized primers, RW01 and DG- 74, were used to amplify a 380-bp fragment that resides within a conserved region of the bacterial 16S rRNA gene and flanked by variable regions V8 and V9. Six bacteria listed in Table 2 were analyzed by using the ABI 377 to obtain their full length of 16S rRNA 380-bp sequence and the genus Lactobacillus was recognized.
To evaluate the antagonistic effect of Lactobacillus isolates against the fresh water fish pathogen, the Aeromonas isolates were isolated from the same trout fishes and identified by culture, morphological and biochemical assays. Evaluation of antagonistic activity of six Lactobacillus isolates against fish pathogen were carried out by well diffusion method.
Among all six isolate with antagonistic effect with A. hydrophila, The isolates F-35 and F-30, known as Lac. acidophilus and Lac. plantarum showed the highest and lowest affect respectively. This report was in agreement with the findings of Joborn who reported inhibitory effect of intestinal bacteria against the growth of Aeromonas hydrophila and Vibrio anguillarum [28].
Similar study had carried out by Chaudhary and Qazi that the antagonistic effect of probiotics assessed on two pathogenic bacteria V. anguillarum and Sphingomonas sp. They observed antagonistic efficacy of nine from twenty probiotic isolates by cross streaking method and amazing results of six isolates by disc and well diffusion methods [29]. It has widely accepted that use of microbial probiotics as echo friend organisms, promote health maintenance and disease prevention and control in aquaculture [16].
Many studies had revealed the antagonistic effect of Lactobacillus as in study carried out by Dhanasekaran et al, they demonstrated that antagonistic outcome of Lactobacillus was responsible for inhibition of Aeromonas population growth in cat fish (Clarias orientalis) [30].
In a study Sica et al, examined in vitro adhesion of twelve isolates of LAB from fish to mucus, and cell surface of rainbow trout and exclusion in compare with salmonid pathogens, Yersinia ruckeri and Aeromonas salmonicida. They observed that all strains were capable to attach to rainbow trout skin and mucus [104-106 cells/cm 2], to glass [104-105 cells/cm2] and to stainless steel [103-104 cells/cm2]. Also sixty percent of LAB strains were able to compete with and successfully excluded Y. ruckeri and all strains were able to displace with it. Besides 75% of LAB strains competed successfully with A. salmonicida and 50% of LAB were able to displace whereas 60% could excluded this pathogen [31].
All documented results indicates that Lactobacillus species are useful bacteria for supporting and health maintenance in host and they are able to change intestinal microorganisms. They can be used as alternative to antibiotics with no serious side effects. Probiotics also maintain gut physiologic balance and don’t disturb it’s homeostasis. Furthermore probiotics are valid alternatives for prophylaxis and can replace with antibiotics and antibacterial agents [32].
Amazingly, probiotics are able to stimulate specific and non-specific immune systems and also gut immune system in fish by increasing immunoglobulin cells and acidophilic granulocytes [33]. Advantages of probiotics are not restricted to stimulation of immune system, they also improve the water quality and nutrition which leads to larva survival and increase aquaculture output [15,34].
Dietary administration of probiotics stimulates natural immune system, probably by adhering and colonization in gastrointestinal tract which leads to increased level of antibodies [35]. There is an increasing interest for use of probiotics in controlling disease in aquaculture because the bacterial resistance to antibiotics is a great concern. World Health Organization, Food and Agriculture Organization has defined the prbiotics as “live microorganism” which can guarantee host’s health if adequately be used [2,36].
Conclusion
Aquacultures produce almost forty percent of world aquatic products while the demand for sea foods is
increasing worldwide. Safety of sea food is a major concern and high consumption of therapeutic and
chemical agents in aquaculture leads to find the alternatives for disease control. Besides the benefits of
antibiotics in health of aquaculture animals, alteration of intestinal microbes and bacterial resistance which
has resulted from chemotherapy, restrict their usage.
Probiotics open a new era in health management strategy for fish gaining with potential advantage of this approach and maintenance of intestinal balance, exerting numerous beneficial effects in host can be considered new perspective in aquaculture farming.
Conflict of Interests
The authors declare that they do not have any conflict of interest.
Bibliography

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!