Biography
Interests
Sabiha Habib & Ambreen Ahmed*
Department of Botany, University of the Punjab, Quaid-e-Azam Campus, Lahore 54590, Pakistan
*Correspondence to: Dr. Ambreen Ahmed, Department of Botany, University of the Punjab, Quaide- Azam Campus, Lahore 54590, Pakistan.
Copyright © 2018 Dr. Ambreen Ahmed, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Bacteria are ubiquitous and can be located everywhere on the planet. The quantification of bacteria and their growth pattern studies in different biological samples is an essential part of microbiology and being used in various researches. Quantifying bacterial evaluation has particular importance in food, water or clinical samples. A variety of techniques have been introduced to determine the viable bacterial counts in sample cultures however CFU count by serial dilution method is the most applied and error-free methodology. Altogether, there is a correlation between bacterial quantification and optical density of bacterial cultures. The aim of the present work is to provide a very simple and cost-effective scientific approach to count bacterial colonies of selected bacterial isolates [Bacillus pumulis (ALa), Bacillus atrophaeus (BL2), Staphyllococcus lentus (E3), Bacillus cereus (AR), T2aii and W6ii] and to correlate with OD600 measurements of those bacterial cultures indicating the efficient and competitive bacterial strains in respect of their growth pattern and rapid multiplication.
Introduction
The microbial creatures in all habitats have been widely studied for many centuries. These are ubiquitously
present in soil, air and water. Among all microorganisms, bacteria constitute a major fraction of microbes. They
exist in many shapes and live as parasites or may have mutualistic relationship with other organisms. Bacterial
growth phases include four phases namely lag phase, log phase, stationary and death phase [1]. Bacterial
enumeration is a significant technical approach for various microbiological studies but it is very laborious
to count bacterial colonies accurately [2]. Several methods have been introduced for the quantification
of bacteria including quantitative PCR, genome probing or fluorescent labelling by following specific
protocols and equipments which may not be present in standard microbiological research laboratories, also
these techniques quantify all bacterial cells including dead or nonviable cells [3-6]. Putman and coworkers
reported a bacterial counting approach by using ProtoCol software but it is a very complex technique as
staining of colonies is required in this method with some dyes to differentiate and contrast between colonies
[2]. In 2012, Brugger and colleagues presented an automated colony counting technique in which CFU
were counted on blood agar plates without staining bacterial colonies but still there was a difficulty about the
identification of dust and abrasions on agar plates [7]. Salazar-Martinez and colleagues used colony forming
units as an indicator of cardiovascular risks in children and adults so CFU count method is important for not
only microbiology but also for medical or clinical purposes [8]. In 2008, Sieuwerts and colleagues reported
a method of bacterial counting where drops of bacterial diluted cultures were spotted on agar plates and
resulting minicolony spots were imaged with a digital camera and quantified through imageJ program. They
used Saccharomyces cerevisiae and Eschericha coli for comparing minicolony method with other conventional
method and observed no significant difference in the results obtained from these methods [6].
Bacterial enumeration through colony forming unit (CFU) is an easy and simple method which is well suited for the assessment of only viable bacteria. CFU basically represent the number of viable bacterial colonies, originated from only one bacterial cell which has the ability to produce a colony. This method has two significant advantages; the first is that it has the capacity to count any bacterial number using several dilutions, too many or too low and second advantage is that it counts only viable bacteria having a particular genotype and exclude debris and dead bacteria. In this method, many bacterial cells aggregate in a single colony so the counts are represented in CFU/ml not in bacteria/ml [9]. It has been reported that each bacterial colony is formed from only single bacterium (colony forming unit). This method is complex and time-consuming. There is an inclination to observe CFU count only for high dilutions as they surely have fewer colonies which are easy to be counted [7]. In this technique, a small volume of bacterial suspension is spread over the agar plate and after incubation, discrete bacterial colonies are formed which are evenly distributed over the surface of growth medium. Total number of bacterial colonies are counted and used to calculated number of bacteria in given volume of bacterial culture [10]. The accurate counting of bacterial colonies is error prone at small dilution due to overcrowding of bacterial populations so high attention is needed to count CFU. For that reason, only a part of plate is observed and used to estimate all CFU counts in corresponding plate after extrapolation [11]. Another method for quantifying the concentration of bacterial cultures includes measuring the optical density at 600 nm (OD600) but this assumption is not as suitable as quantification of bacteria through CFU count as optical density measures all viable and nonviable cells in sample cultures but there is a correlation between CFU/ml and OD600 measurements [12].
In the current study, six previously isolated and characterized bacterial strains (Bacillus pumulis (ALa), Bacillus atrophaeus (BL2), Staphyllococcus lentus (E3), Bacillus cereus (AR), T2aii and W6ii) were used for the quantification of bacteria by plating diluted bacterial cultures on agar plates having different optical densities. Bacterial quantification for each bacterial strain was correlated with optical density and a standard was proposed about number of bacteria at a specific optical density of diluted cultures at 600nm.
Materials and Methods
For quantification of bacteria, six previously isolated and identified bacterial strains Bacillus pumulis (ALa),
Bacillus atrophaeus (BL2), Staphyllococcus lentus (E3), Bacillus cereus (AR), T2aii and W6ii by Fatima and
Ahmed [13]; Jamil [14] and Aslam [15]) were used for the proposed study and were inoculated in liquid
broth media and incubated for 24 hours at 37°C.
Bacterial cultures were serially diluted 10-fold to quantify CFU/mL. 100 μL aliquots from various dilutions
were inoculated on respective labeled agar plates and incubated at 37°C for 24 hours until colonies were
visible. Altogether, OD600 measurements were also recorded for each diluted bacterial culture. After
incubation, the bacterial colonies were counted for all diluted cultures and adjusted according to the dilution
factor. To calculate the number of bacteria/ml, following formula was used:
Number of bacteria/ml = Number of colony forming units/ Volume plated (ml) x dilution used
Results
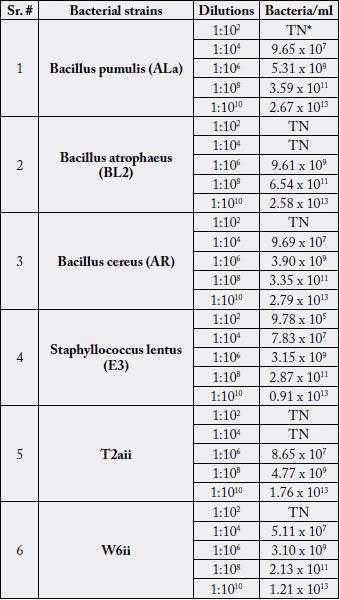
In the current study, bacterial strains i.e., Bacillus pumulis (ALa), Bacillus atrophaeus (BL2), Bacillus cereus
(AR), Staphyllococcus lentus (E3), T2aii and W6ii were considered to examine the number of viable bacteria
in the given volume of respective bacterial culture solutions in different dilutions (1:102, 1:104, 1:106, 1:108,
and 1:1010). Data obtained from CFU/ml quantification were compared with OD600 measurements. At
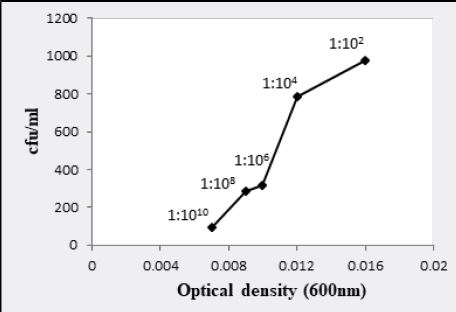
1:102 dilution, bacterial colonies were too numerous to count for all bacterial cultures. At 1:104 dilution,
Bacillus atrophaeus (BL2) and T2aii have shown infinite bacterial colonies while Bacillus cereus (AR) and
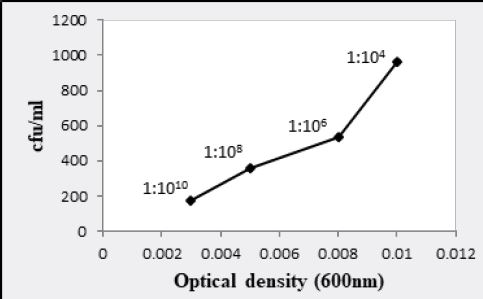
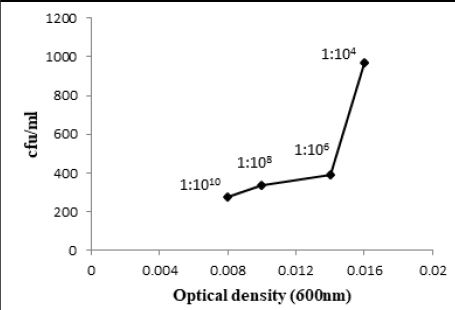
Bacillus pumulis (ALa) exhibited 9.69 x 107 and 9.65 x 107 bacteria/ml having 0.016 and 0.1 optical density
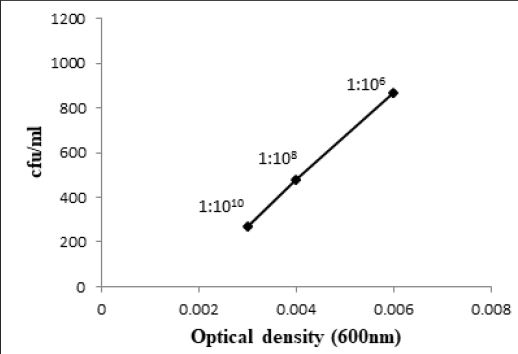
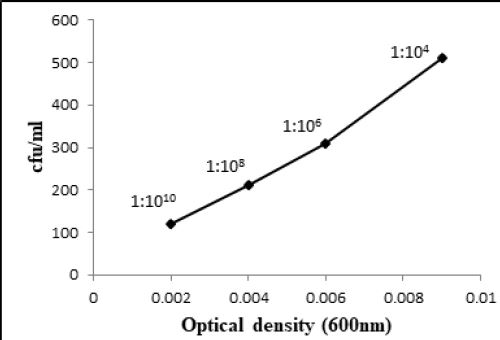
at 600nm respectively (Fig. 3.1 and 3.3). At 1:106 dilution, 9.61 x 109 and 8.65 x 107 bacteria/ml were shown
by Bacillus atrophaeus (BL2) and T2aii cultures respectively having 0.006 optical density for both. At 1:108
dilution, optical density was 0.003 and 6.54 x 1011 bacteria/ml were recorded for Bacillus atrophaeus (BL2)
(Fig. 3.2 and 3.5). In addition, Bacillus cereus (AR), Bacillus pumulis (ALa) and Bacillus atrophaeus (BL2)
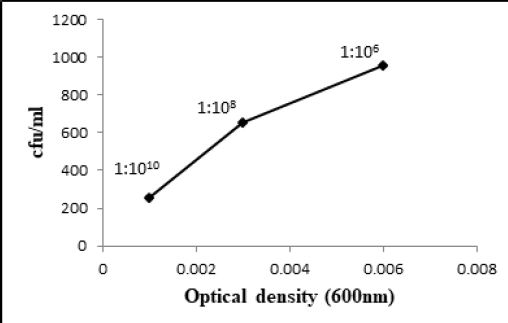
exhibited 2.79 x 1013, 2.67 x 1013 and 2.58 x 1013 bacteria/ml at 1:1010 dilution in the given volume of diluted
bacterial culture having 0.008, 0.003 and 0.001 optical densities respectively (Fig. 3.1, 3.2 and 3.3).
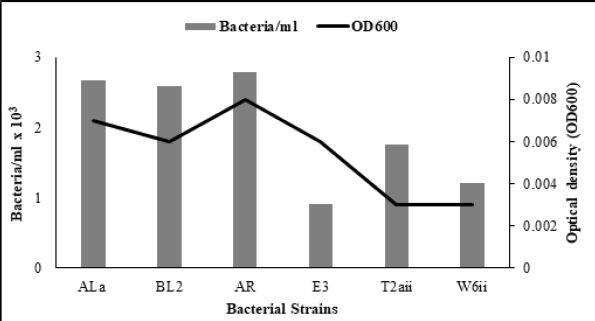
Similarly, W6ii and T2aii have also shown a prominent number of viable bacteria at this dilution upto 2.20 x 1013 and 1.76 x 1013 bacteria/ml while optical densities for these isolates were 0.002 and 0.003 respectively at 600nm (Fig. 3.5 and 3.6). The observations for all bacterial cultures have given significant results of number of bacteria in their respective dilutions having different optical densities (Table 3.1).
*TN = too numerous
Discussion
Quantitative analysis of bacterial colonies is a technical approach in various microbiological researches
providing accurate number of bacteria and information about their growth pattern and level of contamination
in that sample. Every analysis and effect is dependent on the number of bacteria present so correct
enumeration and quantification of bacteria is very necessary. For the estimation of bacterial colonies, several
colony counting approaches are commercially available with high sensitivity and accuracy. These techniques
are designed for qualitative analysis of food and can handle a number of samples having several bacterial
species but are very expensive to use commonly in research laboratories.
The method used in the current study i.e., CFU count method is a very cost-effective and simple technique instead of using high-throughput systems for bacterial counting in sample cultures for which staining or imaging of bacterial colonies are required. Evaluation of bacterial number through CFU count method gives only viable bacterial colonies and also some information about the nature of bacterial growth state like generation time and time of entrance in stationary phase [9]. Quantification of bacterial concentration by OD600 measurements do not distinguish between living and non-living bacteria. Consequently, the measurements do not indicate the exact concentration of rapidly growing bacterial cells [12]. Manually counting of colony forming units was performed at small dilutions having high number of CFU while increasing the dilution of bacterial cultures upto 1:1010 exhibited a decline in colony forming units. At 1:1010 dilution, countable bacterial colonies were recorded for all bacterial cultures i.e. Bacillus pumulis (ALa), Bacillus atrophaeus (BL2), Staphyllococcus lentus (E3), Bacillus cereus (AR), T2aii and W6ii, as the range for countable colonies is 30-300 (Fig. 4.1 and 4.2). In this study, we proposed a standard dilution of all studied bacterial isolates at which bacteria exhibited countable colonies. Altogether, we compared optical density and number of bacteria at different dilutions reflecting a standard of bacterial quantification at a specific OD600 of that bacterial culture.
Our results demonstrate that there is a correlation between CFU/ml and OD600 measurements. OD600 measurements are not quite as accurate, since bacterial cultures used for our study are prone to clump formation in liquid broth but serial dilutions diminished this problem leading to absence of clumps. Different CFU/ml counting and optical densities of each bacteria at the same dilution depends upon the growth pattern, physical properties of bacteria like cell size, light scattering intensity and sedimentation ability of bacterial cultures. This correlation indicates the most competitive and efficient bacteria in respect of bacterial multiplication and rapid growth rate in a specific time period indicating that generation time of such bacteria was very short and maximum bacteria were produced in a very short time span during log or exponential phase before entering into the stationary phase. Data for 1:1010 dilution was considered and observed that diluted culture of Bacillus cereus (AR) presented prominent number of bacteria for 100μl of aliquot and 278.8 CFU/ml while OD600 was 0.008. Although it exhibited maximum bacterial quantification but if it is correlated with optical density then it may indicated that multiplication behavior of Bacillus cereus (AR) was not as good as other bacterial cultures like Bacillus atrophaeus (BL2), T2aii and Bacillus pumulis (ALa) exhibited 258.2, 266.8 and 175.8 CFU/ml even at 0.001, 0.003 and 0.003 OD600 supporting the fact that these bacterial isolates have rapid multiplication rate and excellent growth habit reflecting its competitiveness and efficiency in respect of its division and replication (Fig. 4.3). Number of bacteria/ml as well as optical densities of diluted bacterial cultures were going to be decreased while increasing the dilutions. In various microbiological studies, researchers need to know about accurate number of bacteria in biological samples. In clinical and food analysis, contaminated samples analysis, plant growth promoting studies of bacteria and a number of researches require precise enumeration of bacteria to make their results more significant. In this regard, this study provides a standard about bacterial quantification in selected bacterial strains at different dilutions along with OD600 measurements.
Conclusion
Present work gives an insight towards a very efficient and cost-effective approach of bacterial quantification
by CFU count method and correlation between bacterial counts and OD600 measurements. Our findings
provide a standard dilution at which all bacteria exhibited countable colonies. This study may help in accurate
counting of bacteria in different samples for microbiological and clinical purposes. In addition, it may helpful
in preparing inoculum for plant growth studies by using growth promoting bacteria showing accurate
bacterial counts in bacterial colonization with plants that ultimately involved in growth enhancements.
Acknowledgement
This work is supported by the financial grant from University of the Punjab, Lahore, Pakistan.
Conflict of Interest
The authors declare no conflict of interest.
Bibliography

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!