Biography
Interests
Dr. Shimon Shatzmiller*, Dr. Ludmila Buzhansky, Dr. Rami Krieger, Dr. Inbal Lapidot, Dr. Galina Zats, M. & Dr. Marina Kovaliov
Department of Chemical Sciences, Ariel University, Israel
*Correspondence to: Dr. Shimon Shatzmiller, Department of Chemical Sciences, Ariel University, Israel.
Copyright © 2018 Dr. Shimon Shatzmiller, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Since more than a half of a century it becomes more and more evident: there is a transformation in the microbial world and antibiotic agents are not “magic bullets” any more. The microbes kill thousands in a new pandemic, the nosocomial illness caused by resistant microbes that prosper in the health facilities, mainly the hospital where people look for remedy [1].
“The silent tsunami, crumbling down the pillars upon which modern medicine is built. Antibiotic resistance that turns ordinary disease-causing bacteria into illnesses that can’t be controlled could bring about the “next pandemic,” Centers for Disease Control and Prevention Director Tom Frieden warned at a National Press”
Fighting Superbugs in India: A Short Film [2]
Abstract
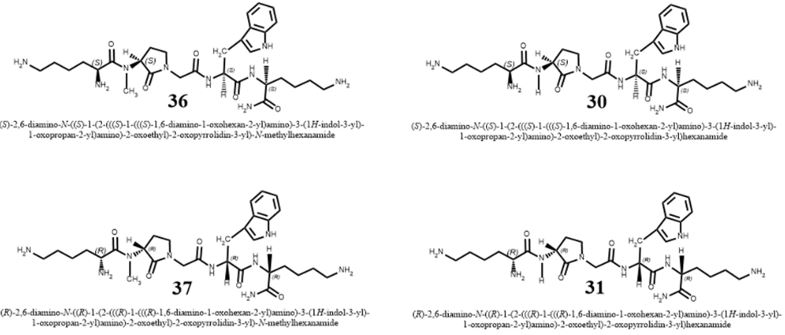
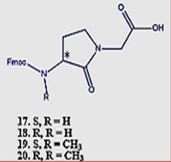
Short surrogates of the pentapeptide K-A-A-A-K, centered on Freidinger’s lactam, were applied in
the eradication of E.Coli (Gram-negative bacteria) and S. Aureus (Gram-positive microbe). It was
found that it was easier to eradicate the gram-positive bacteria, and the eradication was influenced
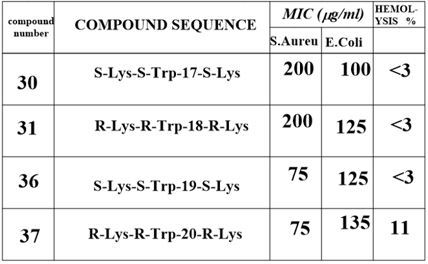
by the N-methylation of the amide nitrogen of 30→31 (table). However, The N-methylated
surrogates were more efficient in the eradication of the E. Coli than the S. Aureus samples. We
were able (Fig. 14) to show that one structural change could make a remarkable difference in the
eradication (MIC) of the different bacteria.
Antimicrobial resistance happens when microorganisms (such as bacteria, fungi, viruses, and parasites) change when they are exposed to antimicrobial drugs (such as antibiotics, antifungals, antivirals, antimalarials, and anthelmintics). Microorganisms that develop antimicrobial resistance are sometimes referred to as “superbugs.”
As a result, the medicines become ineffective, and infections persist in the body, increasing the risk of spread to others.
Introduction
The gut microbes of the Ötzi the Iceman, a 5,300-year-old mummy found frozen in a European glacier in 1991, have shed new light on the history of human migration, scientists say [3]. “We can say now that the waves of migration that brought these African Helicobacter pylori into Europe had not occurred, or at least not occurred in earnest, by the time the Iceman was around.” About half the people on the planet have the bacterium in their stomachs. It can cause ulcers or gastrointestinal distress and is typically spread among children when they play in the dirt.
For as long as the Centers for Disease Control and Prevention (CDC) has measured the prevalence of hospital-acquired infections caused by multidrug-resistant organisms, it has been increasing [4]. According to the CDC, more than 70% of the bacteria now causing hospital acquired infections (HAI) are resistant to at least one of the drugs most commonly used to treat them [5]. It seems important to control these infections because they have been more costly and more deadly than those due to antibiotic-susceptible strains of the same species [6-9]. Nevertheless, despite decades of discussing the control of antibioticresistant nosocomial infections, there has been little evidence of control in most healthcare facilities in most countries. Several articles in this issue of Infection Control and Hospital Epidemiology evaluate the efforts that have been made by hospitals to control them and may help to explain this failure [7].
From early this decade [8], Enterobacteriaceae that produce Klebsiella pneumoniae [9] carbapenemases (KPC) was reported in the USA and subsequently worldwide. These KPC-producing bacteria are predominantly involved in nosocomial and systemic infections [10]; although they are mostly Enterobacteriaceae, they can also be, rarely, Pseudomonas aeruginosa isolates. KPC β lactamases (KPC-1 to KPC-7) confer decreased susceptibility or resistance to virtually all β lactams. Carbapenems (imipenem, meropenem, and ertapenem) may thus become inefficient for treating enterobacterial infections with KPC-producing bacteria, which are, also, resistant to many other non-β-lactam molecules, leaving few available therapeutic options. Detection of KPC-producing bacteria may be difficult based on routine antibiotic susceptibility testing. It is, therefore, crucial to implement efficient infection control measures to limit the spread of these pathogens. One of the most urgent issues in modern healthcare: the global antibiotic resistance pandemic. Current estimates place the annual number of deaths from antibiotic resistant bacteria at around 700,000 worldwide. Unless things change this figure is predicted to rise to 10 million by 2050, with growing numbers of bacteria already fully resistant to every clinical antibiotic available [11].
For as long as the Centers for Disease Control and Prevention (CDC) has measured the prevalence of hospital-acquired infections caused by multidrug-resistant organisms, it has been increasing [12]. Hospitalacquired infections (HAI) have been recognized for over a century as a critical problem affecting the quality of healthcare, and they constitute a major source of adverse healthcare outcomes [13] The emergence of multidrug-resistant bacteria (MDRB) has become a public health problem, creating a new burden on medical care in hospitals, particularly for patients admitted to intensive care units (ICU). Excessive and improper use of broad-spectrum antibiotics, especially in healthcare settings, is elevating nosocomial infections, which not only becomes a big health care problem but also causes great economic and production loss in the community.
According to the CDC, more than 70% of the bacteria now causing hospital-acquired infections are resistant to at least one of the drugs most commonly used to treat them [14].
New strands of pathogenic bacteria cause the nosocomial pandemic all over the world [15,16].

In a recent report published in Lancet Infectious Diseases, scientists discovered colistin-resistant E Coli in 21percent of slaughtered pigs in China. They found isolates in 15 percent of meat sold from those animals in retail sites. They even detected resistant E coli in more than 1percent of hospitalized patients.
Most horrifying, it appears that the resistance is transmitted by plasmids. That means the bacteria don’t just pass on resistance to their “children”; they can pass it among one another and to completely different strains of bacteria. Scientists were also able to detect colistin resistance from the same gene in Klebsiella pneumonia in hospitalized patients. The accompanying editorial to the colistin report called this “a major breach in our last line of defense.”
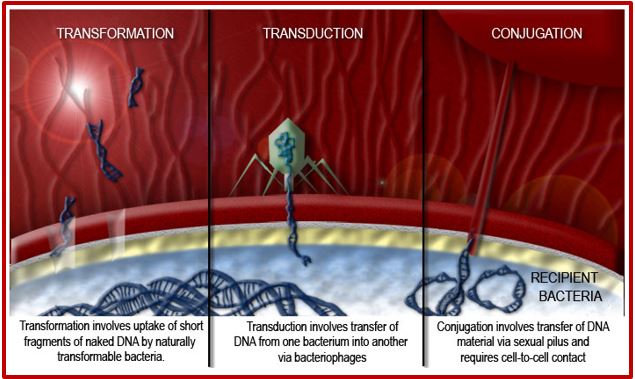
These bugs are mainly the products of genetic transformation that takes place in the process of Horizontal gene transfer, or the process of swapping genetic material between neighboring “contemporary” bacteria, is another means by which resistance can be acquired. Many of the antibiotic resistance genes are carried on plasmids, transposons or integrons that can act as vectors that transfer these genes to other members of the same bacterial species, as well as to bacteria in another genus or species. Horizontal gene transfer may occur via three main mechanisms: transformation, transduction or conjugation.
Transformation involves uptake of short fragments of naked DNA by naturally transformable bacteria. Transduction involves the transfer of DNA from one bacterium into another via bacteriophages. Conjugation involves the transfer of DNA via sexual pilus and requires cell-to-cell contact. DNA fragments that contain resistance genes from resistant donors can then make previously susceptible bacteria express resistance as coded by these newly acquired resistance genes [17].
Let’s look at vertical gene transfer first - how genes are passed from one generation to the next. For both bacteria and humans (and all other animals made of cells), genes are passed down to generations through DNA, the helix-shaped molecules that tell the protein-making machinery in cells what to build. Bacteria, made up of only one cell, have their own DNA and reproduce by dividing into two sister cells, each with the same DNA. In this way, DNA is passed on to the next generation as an almost perfect copy. Even humans started off very much like this single-celled bacterium: When we were but a tiny fertilized egg in our mother’s womb, we were just a single cell that then split into two, then four, then eight cells, each with the same DNA.
While humans then develop specialized cells that then become our trillion-celled bodies, bacteria are singlecell organisms, and their lifecycle is limited to single cells splitting into more single cells, repeatedly.
Each [18] split is a new generation.
Thus, these ‘Superbugs,” or antibiotic-resistant bacteria, have been in the news a lot lately. These types of bacteria can cause infections that are very difficult to treat since they are not killed by conventional antibiotics. While most of them can be eradicated, it requires very powerful (and costly) antibiotics. And most terrifying, we play a role in creating these superbugs. To see how we first need to understand [19] how bacteria reproduce and how they adapt (and share that adaptation to their surrounding buddies). We face the unwanted phenomenon of antibiotic resistance [20].
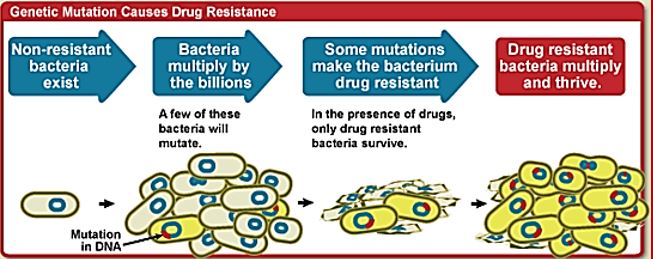
Antimicrobial resistance is the ability of a microorganism to survive and multiply in the presence of an antimicrobial agent that would normally inhibit or kill this kind of organism. Antimicrobial resistance is just one of the many adaptive traits that resilient bacterial subpopulations may possess or acquire, enabling them to out-compete and out-survive their microbial neighbors and overcome host strategies aimed against them. This phenomenon is nearly as old as the discovery of antimicrobials themselves, having been described by pioneers like Ehrlich for trypanosomes8 and Fleming for staphylococci10. What is most alarming today being the rate at which antibiotic resistance often develops and how quickly it spreads across the globe and among different species of bacteria.
Antibiotic Resistance
• Antibiotic resistance is the ability of a microorganism to withstand the effects of an antibiotic.
• It is a specific type of drug resistance.
• Antibiotic resistance evolves naturally via natural selection through random mutation, but it could also be engineered by applying an evolutionary stress on a population.
• Once such a gene is generated, bacteria can then transfer the genetic information, in a horizontal fashion (between individuals) by plasmid exchange.
• If a bacterium carries several resistance genes, it is called multiresistant or, informally, a superbug.
• Causes Antibiotic resistance can also be introduced artificially into a microorganism through transformation protocols.
This can be a useful way of implanting artificial genes into the microorganism.
• Antibiotic resistance is a consequence of evolution via natural selection.
• The antibiotic action is an environmental pressure; those bacteria which have a mutation allowing them to survive will live on to reproduce.
• They will then pass this trait on to their offspring, which will be a fully resistant generation.
One major reason for the proliferation of superbugs is the abuse of antibiotic drugs prescriptions.
When used correctly, antibiotics are lifesaving medicines that fight bacterial infections. Antibiotic misusefor example, taking the drugs unnecessarily, taking the wrong antibiotic, or not taking them exactly as prescribed-creates fertile ground for the growth of antibiotic resistance.
We take antibiotics for granted at our own peril. We assume they will always be available and effective when the truth is that deadly bacteria are becoming immune to existing antibiotics faster than we can find new ones.
At least 80 million antibiotic prescriptions written each year in the United States are unnecessary, which makes improving antibiotic prescribing and use a national priority.
Some superbugs are present in every infection, by abuse of the drugs, the easy to eradicate “normal” bugs are eliminated, but the superbugs remain to further proliferate in the fast pace of minutes for the appearance of the next generation. In this way, the concentration of the “superbugs” - the resistant strands are taking over the bacterial community until there are only “superbugs” in the infection.
Antimicrobial Agent to Fight Bacterial Resistance
Antimicrobial agents have changed human and animal health systematism by revolutionizing our weapons in the war against infectious disease, resulting in improved survivability. There was a time where the perfect antimicrobial drug “The magic bullet a search for the perfect drug” was considered a n achievable target [21]. For both human and animals. However, this health triumph has been tempered by the subsequent realization that bacterial populations can quickly modify them to resist [22].
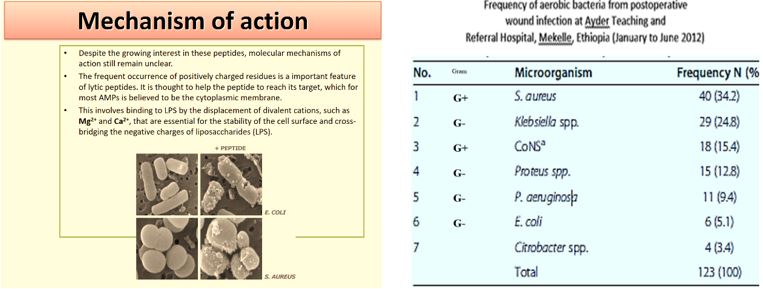
Shai [23] and Matsuzaki [24] showed that antimicrobial peptides could become selective cell agents.
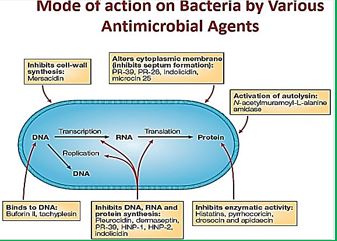
Frequent antibiotic use over long periods of time puts selective pressure on bacteria and causes resistance to spread. When an antibiotic is used to treat a typical bacterial infection, most bacteria are killed. Sometimes, however, a bacterium with an advantage life. This bacterium can then reproduce and pass its advantage on, creating many more antibiotic-resistant bacteria.
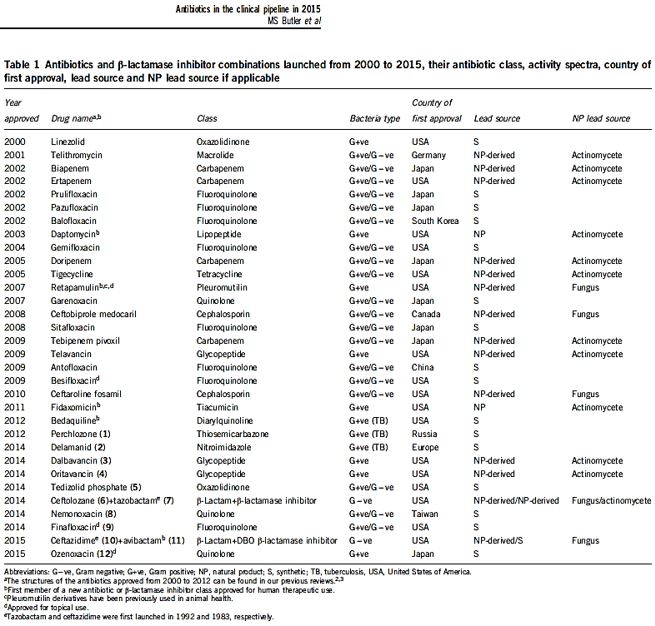
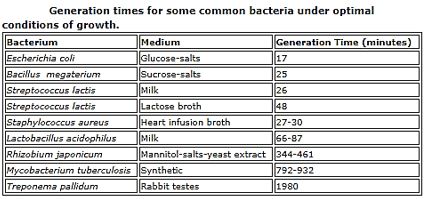
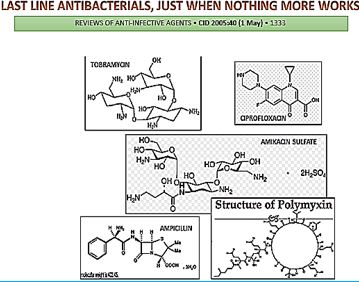
The short list in the table above represents some of the newest antibiotic drugs approved for antiinfection treatment. New compounds are isolated, identified, tested and modified [25], Please notice the poor yields in antibacterial agents in the last decades, only a score of novel drugs was found and approved [26], do not provide weapons to fight infections in healthcare installations (nosocomial infections) and give only little hope to the millions suffering from the fact6 that they do not provide remedy to those that are infected with the resistant microbes and in most cases, must die while treated in hospitals.
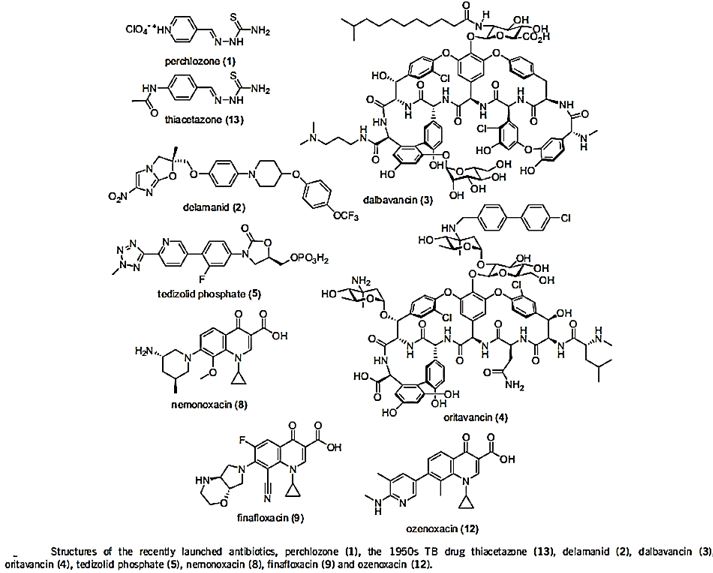
One of the reasons is that even the most modern findings in the area are impotent in the killing of bacteria when resistant strands are the matter.
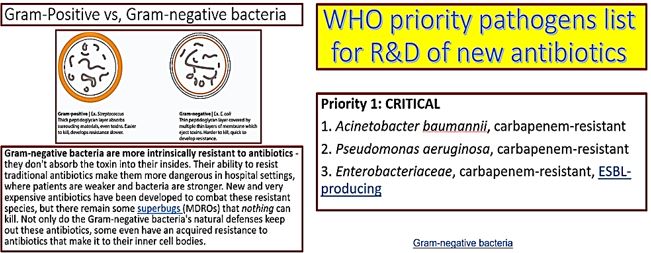
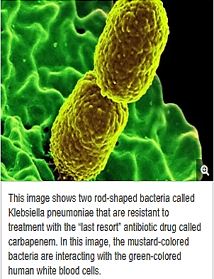
CRE [27] are a type of bacteria that are highly resistant to many antibiotics. They have earned the name “killer bacteria” and “nightmare bacteria” for a good reason. The death rate for those infected with CRE, or carbapenem-resistant Enterobacteriaceae, (The Enterobacteriaceae are a large family of Gram-negative bacteria) is greater than 40 percent, according to the California Department of Public Health (CDPH), and could be as high as 50 percent, according to the Centers for Disease Control (CDC).
From the perspective of drug-resistant organisms, (CRE) is the most serious threat, the most serious challenge we face to patient safety. these (bacteria) are going to greatly impact the kind of surgeries (and) treatments we can have; we’re entering the post-antibiotic era; that’s a very big problem Infections usually occur in hospitals, nursing homes, and other healthcare settings, the CDC said. Members of the general population are generally not at risk.
In March 2016 [28], the CDC reported that “superbugs” - bacteria that are highly resistant to antibiotics - are responsible for one out of seven infections caught in general hospitals, causing an annual estimate of 80,000 in the U.S. Several types of bacterial infections have evolved to ‘deadly invader’ status, and it’s partially our fault. Among the 18 superbugs identified by the CDC are salmonella, drug-resistant tuberculosis, and a staph bacterium known as MRSA (Methicillin-resistant Staphylococcus aureus). These bugs are gaining ‘super’ status because we’ve been careless and stupid with antibiotics like Colistin, rendering them useless, if not dangerous. In other words, we’re helping create deadly types of bacteria. The revival of Colistin is the rehabilitation of a renal [29] damaging agent, it is the best drug to apply when there is no other choice.
The emergence of drug-resistant strains of major pathogenic bacteria such as Staphylococcus aureus is an increasingly serious public health concern [30]. To evade bacterial drug resistance mechanisms, new effective chemotherapeutic agents, which have novel mechanisms of action as well as different cellular targets compared with conventional antibiotics, need to be developed [2]. The shortage of new antibiotics to combat multidrug-resistant (MDR) strains has led to a renewed interest in polymyxins [31]. Polymyxins are active against MDR Gram-negative bacteria such as Pseudomonas aeruginosa, Klebsiella pneumonia, Acinetobacter baumannii, and Enterobacter species [32]. Early reports described high incidences of nephrotoxicity and neurotoxicity during polymyxin therapy [9], and the use of polymyxins was replaced in the 1970s by antibiotics considered to be less toxic. However, recent studies showed that polymyxins have acceptable effectiveness and are considerably less toxic than originally reported. Cationic Colistin (polymyxin B) was re-introduced as “last resort” for treating patients with acute infection based on drugresistant bacteria, However established that the cyclic peptide is very harmful to central organs like heart, liver, kidney brain and is a lethal substance in about 60% of its use causes mortality [33] Polymyxins, a group of polypeptide antibiotics that consists of5 chemically different in 1947. Only polymyxin B and polymyxin E (Colistin) have been used extensively worldwide in Colistin was discovered in 1949 and was non-ribosomally synthesized by Ba5]. Coli of colistimethate sodium in 1959. However, then, due to multidrug-resistant (MDR), gram-negative bacteria in patients with cystic fibrosis. However, the emergence of bacteria resistant to most classes of commercially available antibiotics and the shortage of new antimicrobial agents with activity against gram-negative microorganisms has led to the reconsideration of polymyxins as a valuable therapeutic option.
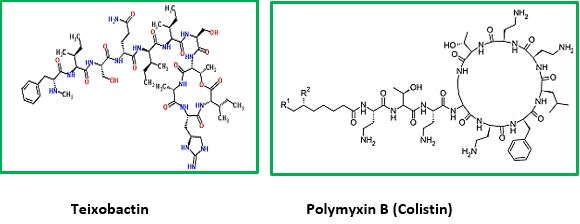
The novel antimicrobial peptide-based Teixobactin identified [34,35]. Recently, Scientists have discovered an antibiotic capable of fighting infections that kill hundreds of thousands of people each year, a breakthrough that could lead to the field’s first major new drug in more than a quarter-century. The new agent that was isolated, identified and synthesized, gives great hope to medicine regarding the combat with resistant bacteria. Some research groups synthesized Teixobactin and some of its analogs. This awakes the hopes that are centered around the following:
1. Selectivity preferred eradication, Gram+ or Gram-.
2. Effective eradication is of all bacteria regular and resistant.
3. Low toxicity to humans.
Not only one target is attacked by Teixobactin, but multiple targets, and they are all lethal. For bacteria, it will be very hard to modify this target, especially this part of the molecule that’s bound by Teixobactin.
Until the discovery of Teixobactin, the following were the top antibiotics in the clinic:
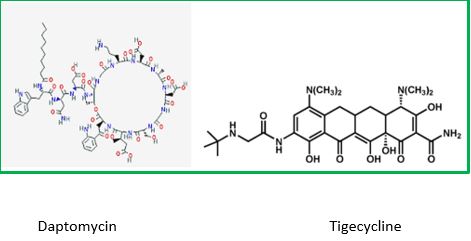
The last major new antibiotic, daptomycin, was discovered in the 1980s by Eli Lilly & Co. After being abandoned in early testing, Antibiotic-resistant bacteria kill at least 700,000 people a year, per a U.K. government review. Unchecked, those infections could lead to 10 million more deaths a year by 2050, the report found.
Tigecycline [36] is the one of newest antibiotic to have been released. It has been on the market for years. It This includes activity against gram-positive, gram-negatives except for P. aeruginosa, many anaerobes, and many atypical pathogens as well as several resistant pathogens. Because it is not vulnerable to many of the resistance mechanisms that affect the beta-lactam antibiotics, it has potential utility in those types of situations. It has been approved for use in complicated skin and skin structure infections, as well as for treatment of complicated intra-abdominal infections due to susceptible pathogens.
These are the only two indications that are approved right now. It has very interesting pharmacokinetics and a long half-life. It will be dosed, after a load, twice a day, and it has a rather low serum concentration, but very good tissue distribution. Tigecycline is available as an intravenous solution. The primary side effects or adverse reactions that have been seen in clinical trials have been gastrointestinal symptoms, usually nausea.
The hunt [37] for new antibiotics is benefiting from recent technological advances which make it easier to rapidly comb the DNA of different soil organisms, which are otherwise hard to raise in a petri dish.
Gonorrhea is the second most common bacterial sexually transmitted infection (STI) in the UK after chlamydia, with almost 35,000 cases reported in England in 2014 [38]. The WHO estimates that 78 million people worldwide contract the disease each year, with most cases affecting young men and women under the age of 25. To control gonorrhea, we need new tools and systems for better prevention. The treatment, earlier diagnosis, and more complete tracking and reporting of new infections, antibiotic use, resistance and treatment failures, Specifically, we need new antibiotics, as well as rapid, accurate, point-of-care diagnostic tests - ideally, ones that can predict which antibiotics will work on that particular infection - and longer term, a vaccine to prevent gonorrhoea.
The soil is a good place to look for new organisms because it is here that bacteria naturally compete for resources and use a range of exotic chemical compounds to kill each other. The paladin chemical, identified by Dr Sean Brady and his laboratory at Rockefeller University, New York, works by attacking a fundamental step in bacterial growth, essentially interfering with a major building block that the bacteria use to build and repair their outer membrane [39].
In their study, published as a letter in the journal Nature Microbiology, the authors cautioned that it is only effective against one group of bacteria - the gram positives, which include MRSA
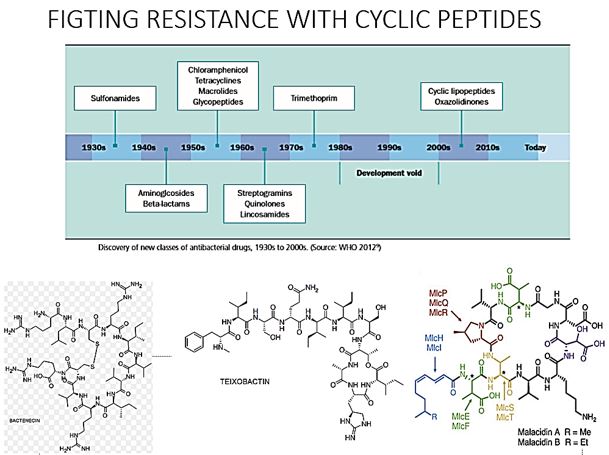
Cyclic peptides are leading the current anti-resistant antimicrobials. Bactenecin, Teixobactin [40], and the Milacainides [41] are the most active that eradicate resistant bacteria strands. The finding new antibiotics to treat gram-positive infections like MRSA was good news but could not address the most pressing need. Our concern is the so-called gram-negative bacteria which are difficult to treat and where resistance is on the increase.
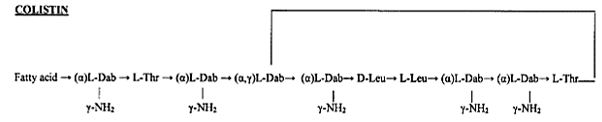
Colistin [42]: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections [43].
Gram-negative bacteria cause pneumonia, blood sepsis [44] and urinary tract infections as skin infections. Twenty-four patients (mean age 44.3 years, mean Acute Physiology and Chronic Health Evaluation II score 20.6) received 26 courses of colistin. Colistin is regarded as one of the last lines of defense against serious diseases, including pneumonia, which cannot be treated by other medicines. Without these drugs, diseases that were commonly treatable in the last century will become deadly once again.
Clinical response was observed for 73% of the treatments. Survival at 30 days was 57.7%. Deterioration in renal [45] function was observed in 14.3% of 21 patients who were not already receiving renal [46] replacement therapy, but in only one case did this deterioration have serious clinical consequences. Colistin was discovered in 1949 and was nonribosomally synthesized by Bacillus polymyxa subspecies colistinus Koyama [47]. Colistin was initially used therapeutically in Japan and in Europe during the 1950s and in the United States in the form of colistimethate sodium in 1959 [6]. However, the intravenous formulations of colistin and polymyxin B were gradually abandoned in most parts of the world in the early 1980s because of the reported high incidence of nephrotoxicity. Subsequently, the intravenous use of colistin was mainly restricted during the past two decades for the treatment of lung infections due to multidrug-resistant (MDR), gram-negative bacteria in patients with cystic fibrosis [10-12]. However, the emergence of bacteria resistant to most classes of commercially available antibiotics and the shortage of new antimicrobial agents with activity against gram-negative microorganisms has led to the reconsideration of polymyxins as a valuable therapeutic option. Currently applied Fosfomycin or Colistin may cause renal damages, it, therefore, demands less damaging antibiotic agents. It seems that we need new antibiotics to treat this class.
Carbapenem-Resistant Enterobacteriaceae (CRE) Infection
CRE infections [48] are most commonly seen in people with exposure to healthcare settings like hospitals and
long-term care facilities, such as skilled nursing facilities, and long-term acute care hospitals. In healthcare
settings, CRE infections occur among sick patients who are receiving treatment for other conditions. Patients
whose care requires devices like ventilators (breathing machines), urinary (bladder) catheters, or intravenous
(vein) catheters, and patients who are taking long courses of certain antibiotics are among those at risk for
CRE infections.
In recent years, carbapenem resistance among Enterobacteriaceae has dramatically increased and represents an important threat to global health.
Some CRE bacteria have become resistant to almost all available antibiotics and can be deadly-one report cites they can contribute to death in up to 50% of patients who become infected. The Therapeutic options for carbapenem-resistant Enterobacteriaceae infections are limited.
Some reports have investigated the clinical efficacy of intravenous Fosfomycin [49] in patients with CRE infections. Michalopoulos et al. reported 11 cases of ICU patients having CRKP infections who were treated with intravenous fosfomycin in combination with other antibiotics; the clinical and microbiological outcome was good with an all-cause hospital mortality of 18.2%, and no adverse events were reported. More recently, Pontikis et al. investigated clinical outcome of 48 fosfomycin-treated (mainly in combination with colistin or tigecycline) ICU patients having XDR fosfomycin-susceptible P. Aeruginosa (n D 17) and K. Pneumoniae (n D 41) carbapenemase-producing isolates; the authors reported a survival rate of 54.2%, evidence of bacterial eradication in 56.3% of cases, and development of fosfomycin-resistance in 3 cases.
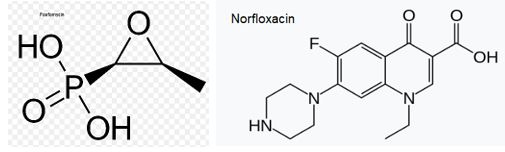
Fosfomycin (also known as phosphomycin or phosphonomycin and the trade names Monurol and Monuril) is a broad-spectrum antibiotic produced by certain Streptomyces species, although chemical synthesis can make it.
As a single dose, fosfomycin is more convenient than multiple-dose therapy Norfloxacin, for the same antibacterial efficacy.
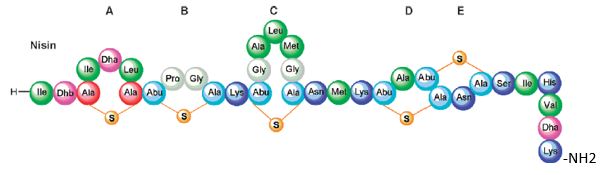
Bioactive Macrocyclic Peptides and Peptide Mimics [50a-c], AIB -based peptide backbone as scaffolds for helical peptide mimics, Antibiotics like Nisin and other peptides and mimics were tested as potent agents for resistant bacteria.
Nisin is a bacteriocin produced by Lactococcus lactis subsp. Latics that exhibits a broad inhibitory spectrum against gram-positive bacteria, including bacterial endospores [51a,b]. Recent studies have shown that the spectrum of nisin activity can be extended to include gram-negative bacteria. Application of nisin in combination with the chelating agent EDTA results in inhibition of Salmonella species and other gramnegative bacteria.
One of the approaches is to learn about the potential application of short peptide mimics like β-turn mimics, on the recognition with perspective to apply this if future drug design. The appearance of β-turns in protein interaction is by far more common than that of other, like γ-turns. Non-covalent [52,53] interactions between the β-turn-mimics and some receptors on the cell wall of bacteria may supply enough energy differences that may allow differentiation between various bacterial transmembrane cell wall receptors [54] due to the receptor and β-turn mimic interactions. The interactions of some β-turn mimics with many classes of proteins which vary in their secondary structure (β-sheets, globular) have been found to rely on the interaction between β-turn mimics and the proteins.
Bacteria Cell Wall and Proteins [55]
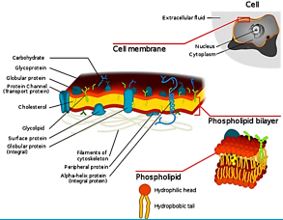
Many variants of β-turn mimic, one count nine β-turn types [56], have been applied so far in this area of research. It has been reported that Surrogate at the β-turn domain of Gramicidin S increase the biological activity [57]. One can read about benzodiazepines, β-turn mimic Hot═Tap for example [58].
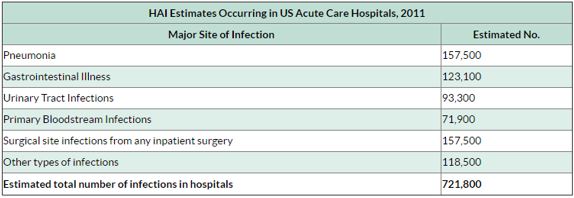
The lack of production and introduction of the newer and effective antibiotic/antibacterial drugs in clinical practice in the post-antibiotic golden age [59] has seen an increase in the emergence of the resistant pathogenic bacterial infections creating a significant problem in the global health of humankind. The situation today is that in 2011, an estimated 722,000 [60] patients contracted an infection during a stay in an acute care hospital in the US; 205 Americans die from hospital-acquired infections (hospital-acquired infections HAI, nosocomial infections [61]) every day.
Compounding the problem of antimicrobial-drug resistance is the immediate threat of a reduction in the discovery and development of new antibiotics [62,63 ].
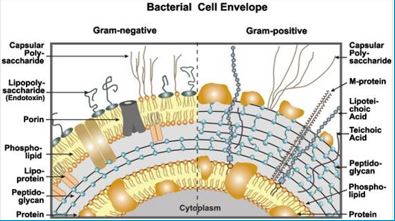
It is now known that many different Gram-positive and Gram-negative pathogens communicate via the production and sensing of small, diffusible signal molecules, to coordinate virulence determinant production [64]. Horizontal genes transfer enables the transfer of resistance from one sort of bacteria to another either by Transformation, involves uptake of short fragments of naked DNA by naturally transformable bacteria. Transduction, involves transfer of DNA from one bacterium into another via bacteriophages. Conjugation which involves transfer of DNA via sexual pilus and requires cell-to-cell contact [65] (see cartoon below). Hunting the nightmare bacteria [66], Therefore, this event, now termed quorum sensing, represents a novel therapeutic target offering the opportunity to attenuate virulence, and thus control infection, by blocking cell-to-cell communication.
Wall [68] teichoic acids are found only in certain Gram-positive bacteria (such as staphylococci, streptococci, lactobacilli, and Bacillus spp); so far, they have not been found in gram-negative organisms. Teichoic acids are polyol phosphate polymers, with either ribitol or glycerol linked by phosphodiester bonds; their structures are illustrated in Figure 2-9. Substituent groups on the polyol chains can include D-alanine (ester linked), N-acetylglucosamine, N-acetylglucosamine, and glucose; the substituent is characteristic for the teichoic acid from a bacterial species and can act as a specific antigenic determinant. Teichoic acids are covalently linked to the peptidoglycan. These highly negatively charged polymers of the bacterial wall can serve as a cation-sequestering [69].
Assessing Compound Permeability in Gram-Negative Bacteria to Enable Rational Drug
Design
In Gram-negative bacteria, the envelope is a sophisticated barrier protecting the cell against external toxic compounds. Membrane transporters, e.g., porins or efflux pumps, are main filters regulating the internal accumulation of various hydrophilic molecules. Regarding bacterial susceptibility towards antibacterial
agents, membrane permeability is part of the early bacterial defense. The bacterium manages the translocation
process, influx and efflux, to control the intracellular concentration of various molecules. Antibiotics and
biocides are substrates of these mechanisms and the continuing emergence of multidrug resistant isolates is a
growing worldwide health concern. Different strategies could be proposed to bypass the bacterial membrane
barrier, comprising influx and efflux mechanisms, in order to restore the activity of antibiotics [70].
Accessing drug targets in the cytoplasm of Gram-negative pathogens is difficult because of the permeability barrier that limits the accumulation of promising antibiotics to effective levels within the cell. The ability to measure compound permeation and accumulation in Gram-negative bacteria is potentially an enabling component of structure-activity-relationships which guide rational drug design and optimization. Strategies addressing these challenges and the development of enabling technologies will be discussed.
While antibiotic resistance is increasing rapidly, drug discovery has proven to be extremely difficult. Antibiotic resistance transforms some bacterial infections into deadly medical conditions. A significant challenge in antibiotic discovery is designing potent molecules that enter Gram-negative bacteria and also avoid active efflux mechanisms. Critical analysis in rational drug design has been hindered by the lack of effective analytical tools to analyze the bacterial membrane permeability of small molecules. We design, fabricate, and characterize the nanofluidic device that actively loads more than 200 single bacterial cells in a nanochannel array. We demonstrate a gigaohm seal between the nanochannel walls and the loaded bacteria, restricting small molecule transport to only occur through the bacterial cell envelope. Quantitation of clindamycin translocation through wild-type and efflux-deficient (ΔtolC) Escherichia coli strains via nanofluidic-interfaced liquid chromatography-mass spectrometry shows higher levels of translocation for wild-type E. Coli than for an efflux-deficient strain. We believe that the assessment of compound permeability in Gram-negative bacteria via the nanofluidic analysis platform will be an impactful tool for compound permeation and efflux studies in bacteria to assist rational antibiotic design.
The introduction of the N-CH3 unit to the Penta peptides surrogates stiffens the structure thereby causes an increase in energy demand for saturation. The fraction of this energy in Gram-negative is smaller than in Gram-positive due to membrane packing composition. It is easier for the N-CH3 to penetrate the outer membrane in gram-positive bacteria and in Gram-negative. Since saturation is achieved in gram-positive with fewer energy demands, the Gram-positive compared to gram-negative are eradicated in preference (through “snorkeling” [71] for example).
The Gram-positive bacteria are easier to penetrate [72] since the outer membrane is poor in membrane proteins. In such circumstance, small energy changes can become significant for the travel of the agent into the outer membrane. The N-CH3 are less flexible and therefore penetrates easier to the membrane. Finally, after the surrogates settle in the inner part of the outer membrane, the Lys ε-amine unit can snorkel [73] out and disintegrate the membranes of both Gram-positive and Gram-negative bacteria. The interactions of an AMP with the membrane cannot be explained by a sequential amino-acid pattern or motif; rather, they originate from a combination of physicochemical and structural features [74] including size, residue composition, overall charge, secondary structure, hydrophobicity and amphiphilic character [75,76].
The interactions of an AMPs surrogates with the membrane cannot be explained only by a particular sequential amino-acid pattern or motif; rather, they originate from a combination of physicochemical and structural features [74] including size, residue composition, overall charge, secondary structure, hydrophobicity and amphiphilic character [75,76]. Also, interactions with the many components that furnish the architecture of the membranes are crucial. From our experiment, we conclude that the venerability of bacteria may depend on small structural variation in the composition of the biocide [8].
Bibliography

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!