Biography
Interests
Ujjal Sen* & Anil Mahadeo Garode
Department of Microbiology, Kalinga University, India
*Correspondence to: Dr. Ujjal Sen, Department of Microbiology, Kalinga University, India.
Copyright © 2018 Dr. Ujjal Sen, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
Yeasts and Fungus are common natural contaminating organisms of frozen buffalo meat at low and freezing environment at abattoir, where smooth colonies recognizes as Yeast (Saccharomyces cereviciae) colonies and hyphae with branching organisms appear as fungus. The aim of the study to isolate yeast and mould, morphology in low temperature and identification through 18S rDNA partial gene sequencing technique. Fungus may belong to class Deuteromycetes, Basidiomycetes or Ascomycetes. Approximately 200 samples of frozen buffalo meat tested and 80% of the sample found positive for fungus and yeast. The experiments exhibits psychrophilic tolerance of yeasts and moulds counts in frozen buffalo meat ranges between maximum 1.48 × 104cfu/gm to minimum 2.72 × 101cfu/gm. 18S rDNA partial sequencing identified internal transcribed spacer gene of fungus as Aspergillus versicolor strain VA 18S ribosomal RNA gene and Aspergillus candidus strain 085241 18S ribosomal RNA gene. The overall study helps to decrease contamination of carcasses during abattoir meat processing environment and create awareness among public regarding fungal and yeast associated food poisoning and food borne disease.
Introduction
Yeast and fungus are most common, contaminating and spoilage organisms associated with bovine slaughter
house low temperature conditions. Slaughter house processing areas differentiated as per temperatures from
room temperature to very low temperature until frozen condition. Carcasses stores at chillers and freezers,
between 0oC - 2oC and between -38oC to -42oC where phychrophilic fungus and yeasts grow well and can spoil the epidermis of meat, cutenious and subcutenious level of buffalo carcasses microbiological testing is mandatory or spoilage may risk potent food borne infection and produce a risk for the consumer. Molds and Yeasts may come from raw materials like buffalo carcasses and by cross - contamination of food handlers [1]. From a study and investigation in present article revealed that in Argentina prevalence of yeast and mold in meat [2].
Food safety depends on adequate manipulation, transportation and storage condition. Some of which such as, yeast and mold can lead to food intoxication and food infection when present in numerous in numbers [3].
Morphological Characteristics
Yeast may be spherical to ovoid, lemon shaped and pear shaped, cylindrical, triangular or even elongated branching into a false or true mycelium. They also differ in its size and their structural visible parts are cell wall which may be metachromatic albuminous or starchy. Special staining is necessary to demonstrate the nucleus. Most yeast reproduces by multilateral or polar budding, a process in which some of the protoplasm bulges out the cell wall. Finally bulge grows in six and matured in a yeast cell. In some of the yeast bud appears to grow from a tube like projection from the mother cell. Nucleus material replicated to form mother cell and daughter cell. A few species of yeasts reproduced by fission and reproduces by a combination of fission and budding. Some yeast species form film or scum on the surface of liquid, are called filamentous yeasts or oxidative yeasts. The lower limit of water activity for ordinary yeast ranges from 25 to 30oC and optimal range of acidity is pH 4 to 4.5. All fermentative yeasts are able to ferment glucose to produce ethanol and carbon - di - oxide. Yeasts can utilize both simple inorganic (ammonia and in some species nitrate) and organic (urea) compound as a nitrogen source, but also utilize amino acids, peptides and polypeptides. In addition to sugars (sources of carbon, oxygen and hydrogen) and N - containing compounds, the yeasts need several other important biological growth factors.
The mold is termed as certain multicellular, filamentous fungi, their growth on buffalo meat is recognized by its fuzzy and cottony appearance. Basically mold act as spoilage organisms in frozen buffalo meat. Molds also used as beneficial purpose: they tend to produce several enzymes and antibiotics. Microscopically molds consists of a mass branching, interwined filaments called hyphae (singular hypha), and the whole mass of this hyphae is known as mycelium. Hyphae may be vegetative or fertile based on biological function. Hyphae function as nutrition of the mold and the fertile ones with the production of reproductive parts. The reproductive parts of structures of molds are the spores, which are mainly asexual.
Such spores are produced in large numbers and are readily spread by air to alight and start new mold growth of plant where conditions are favorable. Molds usually grow in less moisture and water activity for spore germination as low as 0.62 and as high as 0.93 [4].
There are various molds & yeasts are contaminating frozen buffalo meat and those are: Aspergillus, Penicillium, Candida, Rhodotorula. Aspergillus species are A. niger, A. flavous, A. fumigates, A. terreus. Penicillium species are P. chrysogenum, P. expansum, oxalicum, P. citrunum. Aspergillus flavus isolates produced aflatoxins and found toxigenic. Aspergillus ochraceus produced ochratoxins. The rate of proteolytic activity is slightly higher in buffalo meat recovered fungal isolates than cattle. Candida albicans is known as yeast and recovered from both types of meat. Apart from above mentioned genera and species other molds, such as, Cladosporium species, Alternaria species, Helminthosporium species, Paecilomyces species, Mucor species, Rhizopus species, Fusarium species, Geotrichum species are found in frozen buffalo meat and raw meat. In this publication it is reported around 200 meat samples tested from meat and surrounding environment and results shows different yeasts and molds produces aflatoxins, ochratoxins, proteinase and lipase enzymes which contaminate the meat and spoil the meat in Egypt [5]. In recent study, yeast and molds count observed 2.13 × 103cfu/g in buffalo meat slices incorporated in finger millet, oats and chickpea in India [6]. Fungi pollute meat and other animal tissues in the time of slaughter and as result of environmental factors, which favour the growth of fungi and mycotoxins production as: high temperature, water activity or relative humidity, CO2, pH, oxidation potential, and unclean animal houses. A research paper also published that yeast and molds counts present in meat samples collected from non-industrial products from shops in San Luis, Argentina [7]. Most mycotoxins are stable compounds that are not destroyed during food processing or home cooking. Certain foodborne molds and yeasts may also elicit allergic reactions or may cause infections like mycosis. Although most foodborne fungi are not infectious, some species can cause infection, especially in immunocompromised populations, such as the aged and debilitated, HIV - infected individuals, and persons receiving chemotherapy or antibiotic treatment.
Methodologies
Collect 100gm of randomly selected frozen buffalo meat samples from specific cuts or different cuts from the abattoir, transport it into insulated shipping container enough gel - type to maintain refrigerant at 6°C or below. Upon receipt in the laboratory, store the samples at 4°C and analyze it quickly. If analysis cannot be started within 4 hours after collection, freeze the samples promptly and store at - 20°C until examined. Thaw at room temperature and proceed with analysis as usual. Maintain frozen samples at - 20°C until examined. Out of 100gm sample 25gm of frozen buffalo meat sample are weighed and add 225mL of 0.1% peptone water, homogenized the mixture in a blender, afterwards stirrer in screw capped bottle for few seconds or a minute and used as pre - enrichment solution according to FDA, BAM, 2003 sampling method
Make serial dilution from 10-1 to 10-6. With the help of pour plate technique from pre-enrichment, transfer 1mL of pre-enrichment broth to rose bengal chloramphenicol agar or potato dextrose agar plates serially placed from 10-1 to 10-6 as per serial dilution at 40°C - 45°C. Allow the plates to solidify with gentle tilt and stirrer. Incubate the plates at 25°C for 5 - 7 days without disturbing the plates. Probably, if no fungal colonies appear within 5 days, consequently, incubate the plates for another 2 days. Hence, after 7 days fungal colonies may appear on the plates. Observe the presence or absence of fungal & yeast colonies and Count the colonies [8].
Yeasts observed as whitish or colored smooth colonies and molds observed as rough whitish or colored mycelial colonies on rose bengal chloramphenicol agar or potato dextrose agar.
Yeast and molds are observed microscopically by stained with lactophenol cotton blue with oval shaped yeast and molds spores and branching etc.
There are 200 samples of frozen buffalo meat experimented, out of 200 samples 158 samples shows positive results of yeasts and moulds colonies. As per above experiments the tolerance of yeasts and moulds counts ranges from maximum 1.48 × 104cfu/gm to minimum 2.72 × 101cfu/gm. They survive the extreme low temperature during processing at abattoir so, yeasts and moulds also extremely psychrophiles. Hence, it is proved that mycotoxins secreted by fungus can lead to a fatal food borne zoonotic diseases for human being.
Experiments Conducted with Probiotic Organism Yeast Against Other Pathogen Isolated from Meat Microbial Population
Probiotics are live microorganisms thought to be beneficial to the host organisms. The beneficial antagonistic
effects and advantages of antagonistic features of yeast are produce allergy - causing spores and mycotoxins
and synthesize antimicrobial metabolites. Yeast can colonize on dry surface for long time. Yeast is economic
than compare to other chemical antimicrobial because substrate used for growth are cheaper. Use of Yeast as
probiotic agent is convenient because yeast cell has high amount of vitamins, minerals and essential amino
acids.
Some Yeasts are available to synthesize toxin that has protein and glycoprotein which inhibit other strains toxins. These synthesized protein named as killer protein or killer toxin, hence yeast called as killer yeast. In 1963 Killer yeast Saccharomyces cereviciae by Bevan and Makower. Some of other killer strains are Candida, Cryptococcus, Hanseniapora, Hansenula, Pichia etc. Some commercial used as Bio-control product to prevent food deterioration. Certain concentration of yeast give ‘Yield plus’. Such as Aspire, Candida oleophia, Cryptococcus albidus has antagonistic effects [9]. Recent paper updated the knowledge to cultivate cellulytic bacteria from buffalo meat and its lumen and used as probiotic agent against the pathogenic flora [10].
In these experiments, 50μL yeast cell concentration as probiotic agent used against Vibrio cholerae and Salmonella typhimurium in two different tests and observed positive antagonistic effects.
Contamination of Food by Mold and Yeast
Molds are microscopic fungi and can found both in plants and animals. Molds grow in wide range of foods and spoil the food. Some form toxins and can cause illness. Growth of mold in food when its highly acidic and low moisture. Freezing does not destroy the mold. Mold needs air to grow. Most molds produce spores. These spores can be transported by air, water, or insects. Molds form spores which, when dry, float through the air and find suitable conditions where they can start growing again. Molds also tolerate salt and sugar so they can grow in opened jars of jams and jelly and on cured, salty meats, such as ham, bacon, salami, and bologna. Molds also have branches and roots that are like very thin threads. The roots may be difficult to see when the mold is growing on food and may also be very deep in the food. Therefore, if you see mold on a food, you must throw out all of the food and not just the moldy portion. Some molds cause allergic reactions and respiratory problems. And a few molds, in the right conditions, produce mycotoxins (or poisons) that can make you sick [11].
Yeast are another type of fungi and yeast is commonly found on plants, grains, fruits, and other foods
containing sugar. They are present in soil, in the air, on the skin and in the intestines of animals and in some
insects. They are transferred from place to place by people, equipment, or food and air currents. Yeast cause
food to spoil and do not cause foodborne illness.
Different Sources of Yeast & Fungal Contamination in Slaughter House
Buffalo carcasses handled in different kinds of lower temperatures during processing in slaughter houses by the butchers and food handlers. There are immense risk for contamination and cross- contamination during pre and post processing. The risk relies on moisture condition, temperature condition and other environmental condition while handling. Underneath few of the categories discussed regarding frozen meat contamination and spoilage. The air quality in slaughter house with different temperatures from chilling to frozen (25 - 30°C, 8 - 10°C, 0 - 2°C, - 38 to - 42°C, - 18 to - 20°C) are checked with total 250 samples and exhibit the presence of yeast & mould which can cause spoilage of buffalo meat at chilling & frozen temperature too. In slaughter house water also a very effective source of initiation of contamination as per its utilization. The research and more risk of microbiological assessment in buffalo slaughter house carried out by direct experiment of swab of food handlers and butchers, even used instruments and equipments according to the international standards and set microbiological limits.
Molecular Species & Strain Identification of Fungal Isolates Through 18s rDNA Sequencing Technique
DNA sequencing is the process of determining the sequence of nucleotide bases (As, Ts, Cs, and Gs) in a
piece of DNA. In recent times, with the right equipment and materials, sequencing a short piece of DNA
is relatively straight forward and sequencing an entire genome (whole organism’s DNA) remains a complex
process. It requires breaking the DNA of the genome into many smaller pieces, sequencing the pieces, and
assembling the sequences into a single long “consensus sequences.” However, thanks to new methods that
have been developed over the past two decades, genome sequencing is now much faster and less expensive
than compare to Human Genome Project start superscript & end superscript. For an example, Regions of
DNA up to about 900900900900 base pairs in length are routinely sequenced using a method called Sanger
sequencing or the chain termination method. Sanger sequencing was developed by the British biochemist
Fred Sanger and his colleagues in 1977. Genomes are now typically sequenced using other methods that
are faster and less expensive. Sanger sequencing is still in wide use for the sequencing of individual pieces
of DNA, such as fragments used in DNA cloning or generated through polymerase chain reaction (PCR).
Sanger sequencing is similar to DNA Replication in an organism. Sanger sequencing contain ingredients
like a DNA polymerase enzyme, a primer – which is short piece of DNA that binds to the template DNA
& acts as a starter for the polymerase, four DNA nucleotides (dATP, dTTP, dCTP, dGTP), the template
DNA to be sequenced & Dideoxy or chain-terminating, versions of all four nucleotides (ddATP, ddTTP,
ddCTP, ddGTP), each labeled with a different color of dye.
The DNA sample to be sequenced is combined in a tube with primer, DNA polymerase and DNA nucleotides (dATP, dTTP, dGTP, and dCTP)107. The four dye-labeled, chain-terminating dideoxy nucleotides are added as well, but in much smaller amounts than the ordinary nucleotides.
The mixture is first heated to denature the template DNA (separate the strands), then cooled so that the primer can bind to the single-stranded template. Once the primer has bound, the temperature is raised again, allowing DNA polymerase to synthesize new DNA starting from the primer. DNA polymerase will continue adding nucleotides to the chain until it happens to add a dideoxy nucleotide instead of a normal one. At that point, no further nucleotides can be added, so the strand will end with the dideoxy nucleotide. This process is repeated in a number of cycles. By the time the cycling is complete, it’s virtually guaranteed that a dideoxy nucleotide will have been incorporated at every single position of the target DNA in at least one reaction. That is, the tube will contain fragments of different lengths, ending at each of the nucleotide positions in the original DNA. The ends of the fragments will be labeled with dyes that indicate their final nucleotide. After the reaction is done, the fragments are run through a long, thin tube containing a gel matrix in a process called capillary gel electrophoresis. Short fragments move quickly through the pores of the gel, while long fragments move more slowly. As each fragment crosses the “finish line” at the end of the tube, it’s illuminated by a laser, allowing the attached dye to be detected. The smallest fragment (ending just one nucleotide after the primer) crosses the finish line first, followed by the next-smallest fragment (ending two nucleotides after the primer), and so forth. Thus, from the colors of dyes registered one after another on the detector, the sequence of the original piece of DNA can be built up one nucleotide at a time. The data recorded by the detector consist of a series of peaks in fluorescence intensity, will be seen in chromatogram. The DNA sequence is read from the peaks in the chromatogram [12-16].
The 18S rDNA Sequencing technology explains and involves (DNA extraction, PCR amplification, Gel verification, Purification, Sequencing, Sequence assembling, Bioinformatics analysis (BLAST in Various Databases) and Construction of Phylogenetic tree. Here in fungal 28S rRNA different Region Sequencing is used along with different ITS region primer sequencing for the better identification of species and strain of fungus. As reported in a journal it is believed that 18S rDNA approach are useful tool for screening of fungal communities and represent more comprehensive picture of the community than plate culturing, also demonstrate fungal biodiversity within particular environment [17]. In another study reveals, 4 fungal samples from soil utilized for 18S rDNA and internal transcribed spacer (ITS) polymerase chain reaction (PCR) pairs were tested for specific towards target fungal DNA to know their biodiversity [18]. Another study on fungal identification by rDNA sequence analysis to explain the role of microbial community in vermicomposting exhibit internal transcribed spacer is a special sequence that present in between sequence of ribosomal DNA which present in multiple copies and act as a consecutive sequences. Present study of fungi carried out by PCR technique with specific primer. The length of the primer different for different group of fungi [19].
In this article present research experiments revealed from buffalo meat two different isolates of fungi identified microscopically and for species with strain level identification through 18S rDNA sequencing carried out and results of sequencing with chromatogram and blast results underneath given elaborately:
TTTTTTTTTTTTTTTCTTCCCGCCGCNCTGCTGTTTTTTTTTGGGGGGGTTTCCCCCCC
CTCTTTTGGGGTTTTCCCCGGGGCCCGTGGGGTTGGGGGGCTTTTGGGCGGCCCTT
GGGCGCCCCTTGGTTCGGGCGTCCCCCCCCCCCCTTTCCCCCGNGGGGCCGGCTGG
GGGCTGGTGGGGCCTTTGGCCCCTTTCCCCCCGGGGGGGGGGGCCCCCCCCCCCCC
CCGGCCGGGCTTGTGGGCTGGTTCGCTGGGGGGGGCTGCCCCCCCGGGTGGCCCG
GGCCCCCTTTGCTTTCCGGGGCCCGGGGGTGTCCCTACTTTTTGCCTTCCCCCTTTCC
TTCCCGCGTTCCGGTGCTTTTTTCCCTGGGGCGGGGGCCCTNGCGTTCCCTTTTGCCC
GTTTTGCTTGTTTTGTTNTTCCGGCTCCGGNCTGCCTCCTCTCTCGGGCCGCCNGTTC
GGTCGTCCCCCGCGGCTGGCCCCCCCGGGGGTTNGCCCGCCGCGGCCCNCGNNTTN
NGGGNNNGCCGGGGGGGGGTNGGGCGCCNGGNGGCGGCCNCCNNNNCNGGNNN
TGCTTTTTTTTTGGGGGGGGGCTGCGGAAGGATCACCGGGCGCCCACCTCCACCCGT
GAATATTACTGAGTGCGGGCTGCCTCCGGGCGCCCAACCTTCCCACCCGTGAATACC
TAACACTGTTGCTTCGGCGGGGAACCCCCTCGGGGGCGAGCCGCCGGGCCTAACAC
TGTTGCTTCGGCGGGGAACCCCCTCGGGGGCGAGCCGCCGGGGACTACTGAACTTC
ATGCCTGAGAGTGATGCAGTCTGAGTCTGAATATAAGACTACTGAACTTCATGCCTGA
GAGTGATGCAGTCTGAGTCTGAATATAAAATCAGTCAAAACTTTCAACAATGGATCTC
TTGGTTCCGGCATCGATGAAAATCAGTCAAAACTTTCAACAATGGATCTCTTGGTTCC
GGCATCGATGAAGAACGCAGCGAACTGCGATAAGTAATGTGAATTGCAGAATTCAGT
GAATCGAACGCAGCGAACTGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCAT
CGAGTCTTTGAACGCACATTGCGCCCCCTGGCATTCCGGGGGGCATGCATCGAGTCT
TTGAACGCACATTGCGCCCCCTGGCATTCCGGGGGGCATGCCTGTCCGAGCGTCATT
GCTGCCCATCAAGCCCGGCTTGTGTGTTGGGTCGCTGTCCGAGCGTCATTGCTGCCC
ATCAAGCCCGGCTTGTGTGTTGGGTCGTCGTCCCCCCCGGGGGACGGGCCCGAAAG
GCAGCGGCGGCACCGTGTCCGTCGTCCCCCCCGGGGGACGGGCCCGAAAGGCAGC
GGCGGCACCGTGTCCGGTCCTCGAGCGTATGGGGCTTTGTCACCCGCTCGACTAGG
GCCGGCCGGGGTCCTCGAGCGTATGGGGCTTTGTCACCCGCTCGACTAGGGCCGGC
CGGGCGCCAGCCGACGTCTCCAACCATTTTTCTTCAGGTTGACCTCGGATCAGGCGC
CAGCCGACGTCTCCAACCCATAGGGATACCCGCTGAACTTAAGCATATAAANACCCG
GAGGAGGAAATCATTACNGAGTGCGGGCTGCCTCCNGGCGCCCAACCTCCACCGTG
NANATACTNNAAATGNTTGCTTCGGCGGGGAAAACCCCTGGGGGGCGAGCCGCGG
GGCACTAAGGAACTTCAGGCTGGAGAGTGATGCACTCTGAGTCTGAATATAANATCA
GTCAAACTTTCACAATGGATCTNTGGGTTCCGCNTCGATGAAGAAGCAGCCAATGCG
ATNAGTAATGTGAATTGCAAAAATCATGAAACATCAGGTCTTTTACGCAATAGGGCCC
GGGCAATCGGGGGGGCAGCCGGTCTAAGCTCATGCTGCCCAAGCCGGGCTGGGGTG
TTGGGGTTGCGCTCCCCCCGGGGAAGGGGCCCAAAGGCAGCGCCGGCCCCTGTCTG
GCCTCTTGCGTAAAGGGGGTTTTTACCCCTTTACAAGGGGGGGCCGGGCCCCGCCCA
ACAACCCCCACCCTTTTTTTCTCTTTTAACCCGAAACAGGGGGGGTACCCCTTAATACA
TAACCCCCGGAGAAAGAAAAAAAGAAAAANNNNNNNNNCNCNCCCCCCCNGGNGG
GGGGNGCCGCCACAAAACAACCCCCCCCACCCCANAACACCAAAGGGGANGAGTAA
GAAAAAAACACTTTTTAAAAAAAAAAATTGTGGTGGTGGGGGGGCCTTGCTCCGCGC
GCTGCCCGGGGGGGGGGGGGAAAGAAGAGGAGCCCGCCCCCCCACCTCCCCCCCTG
GGGGGGGGGGGGGGGGGGAGGAGGGGAAGGCACGCGCCCCCGGGGCCCCGGGG
GGGGGGGGGGAAGAGAAAAAAAAAAACCCAAAAACAAAGAAAAAAAAAAAANTAT
ATTAATTTTTATTATTACTTACTATCCTTGGGGAATCGAGCGCGGGCGGCCGCGGGGG
GNCG.
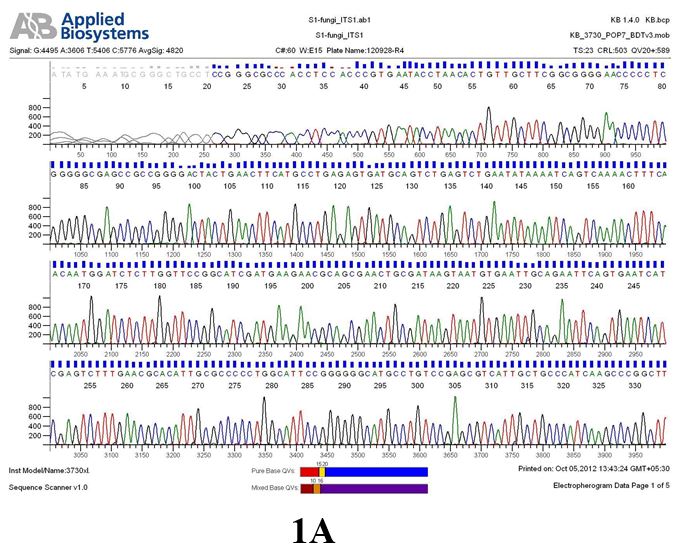
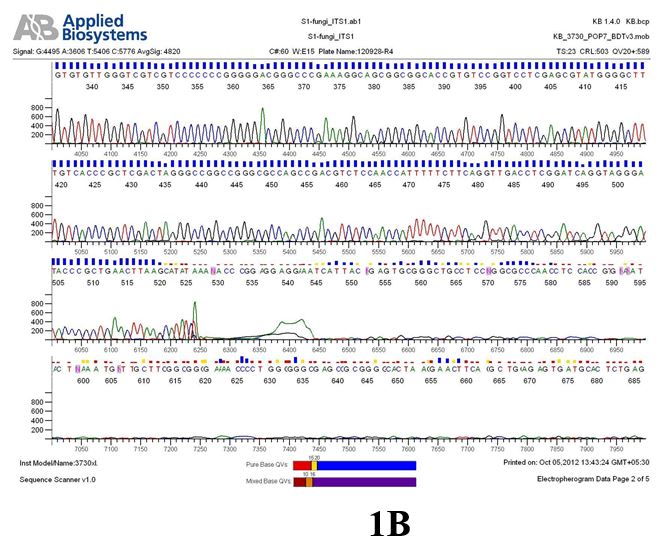
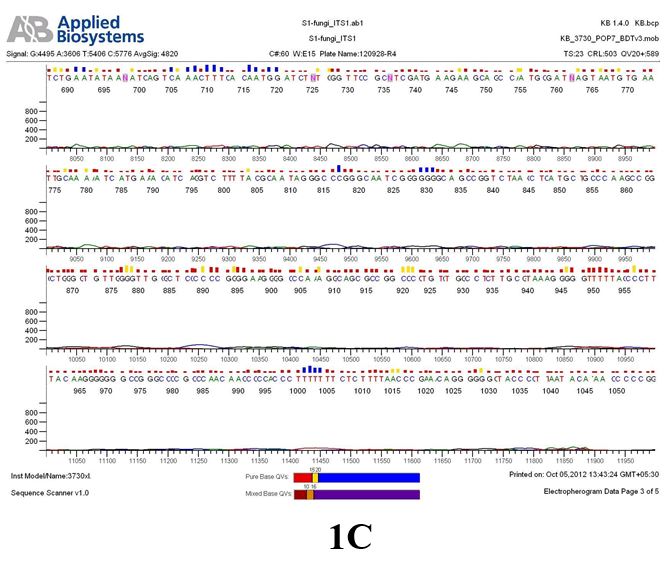
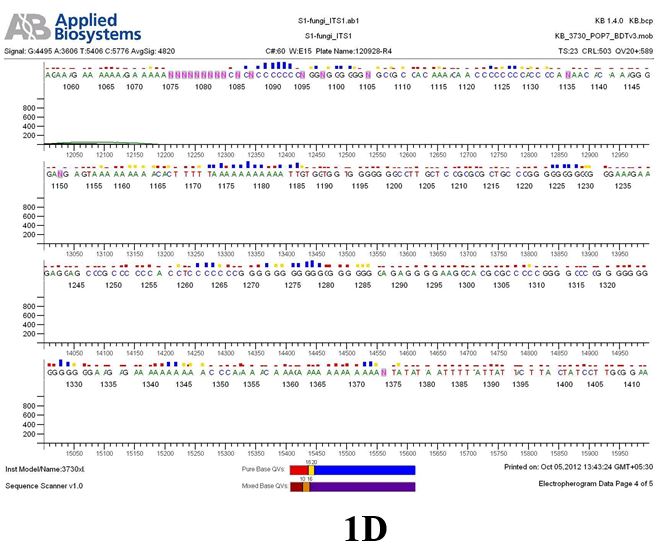
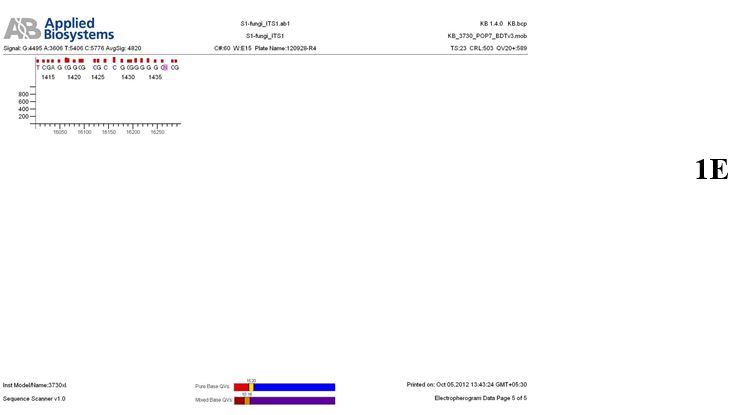
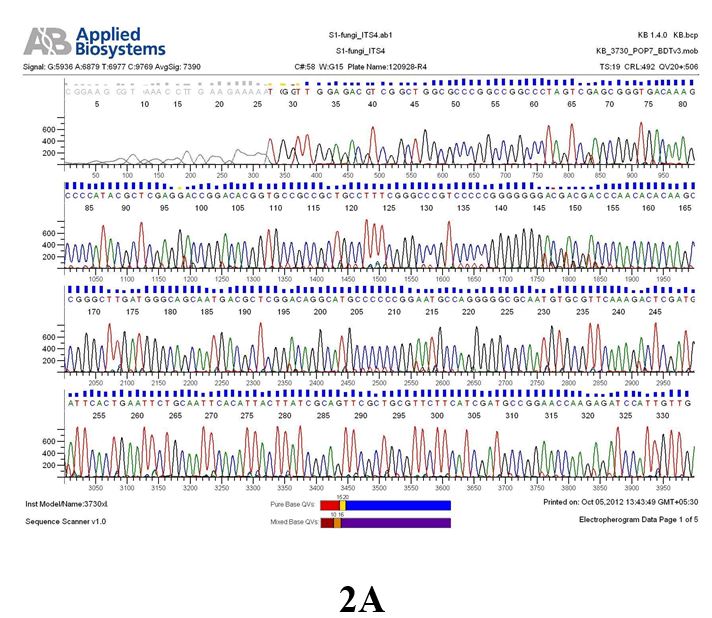
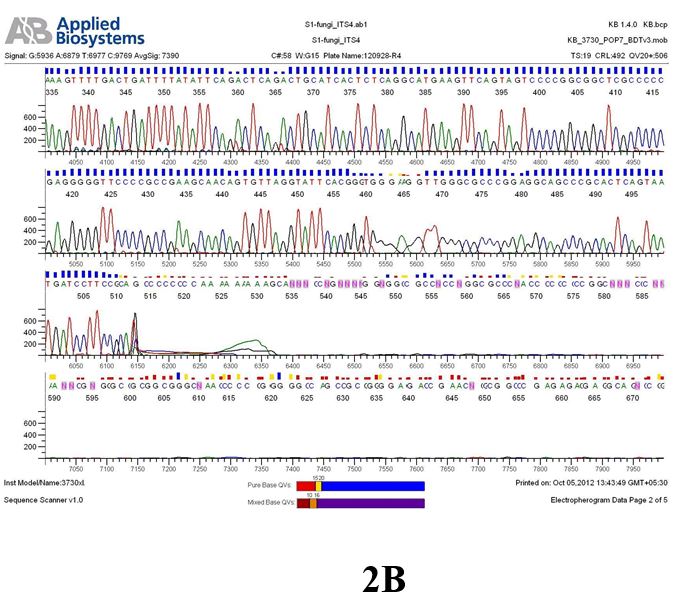
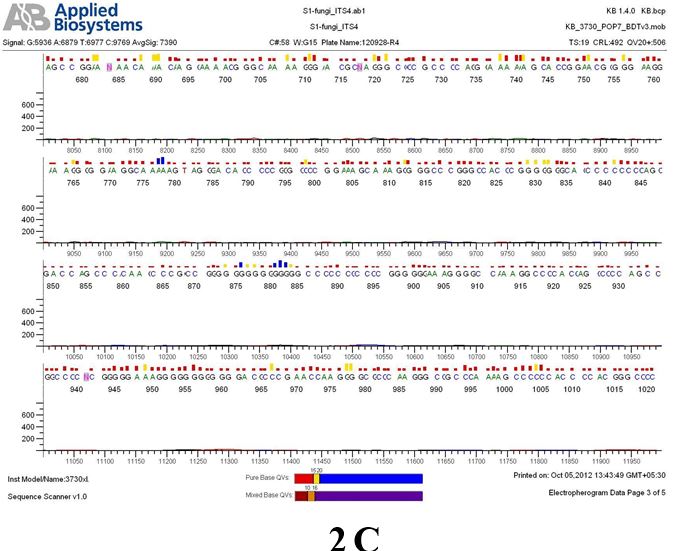
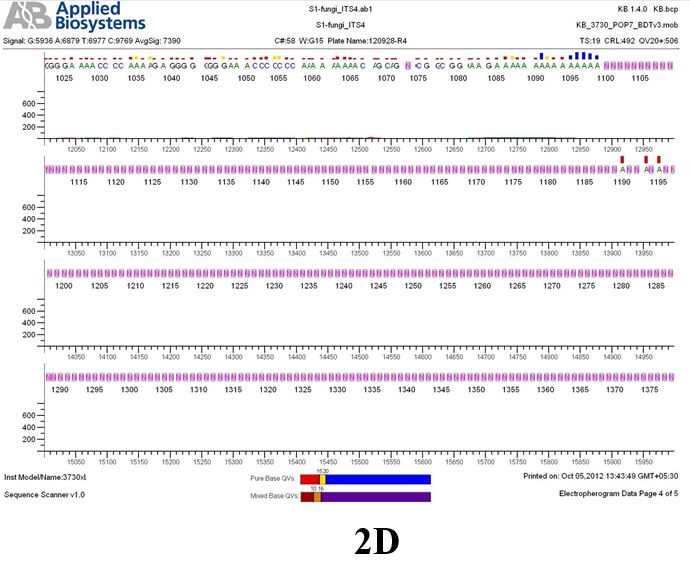
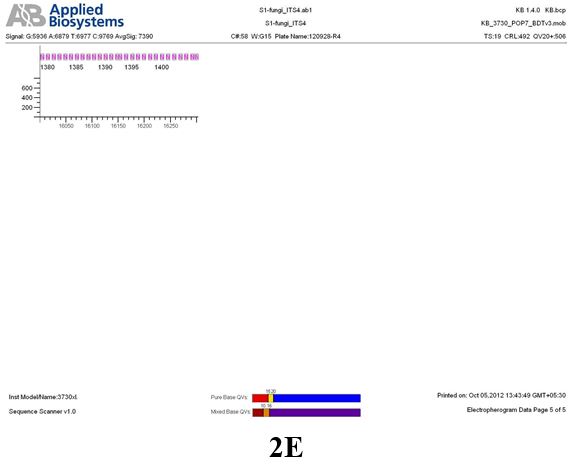
Result
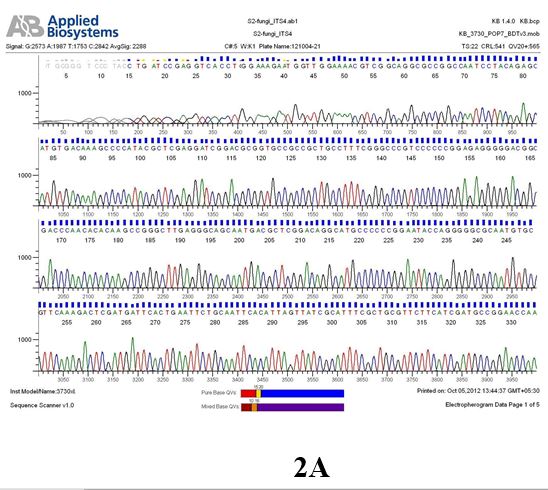
The above sample of filamentous fungal culture DNA has been amplified in PCR. Hence, observed it’s sequencing pattern in forward, reverse and consensus sequences mention the pattern above. The quality of the sequencing results observed through chromatogram designed with two different internal transcribed spacer (ITS) polymerase chain reaction primers named as S1_fungi_ITS1.ab1 and S1_fungi_ITS4.ab1, both pairs with fungal ribosomal DNA which shows picks with four different colors position on accurate base pairs on chromatogram in forward and reverse direction. The sample S1 is identified to be Aspergillus versicolor strain VA 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence.
Sequences S2
CCTTTGGGCCCACCTCCCATCCGTGTCTATTGTACCCTGTTGCTTCGGCGGGCCCGCC
GCTTGTCGGCCGCCGGGGGGGCGCCTCTGCCCCCCGGGCCCGTGCCCGCCGGAGAC
CCCAACACGAACACTGTCTGAAAGCGTGCAGTCTGAGTTGATTGAATGCAATCAGTTA
AAACTTTCAACAATGGATCTCTTGGTTCCGGCATCGATGAAGAACGCAGCGAAATGCG
ATAACTAATGTGAATTGCAGAATTCAGTGAATCATCGAGTCTTTGAACGCACATTGCGC
CCCCTGGTATTCCGGGGGGCATGCCTGTCCGAGCGTCATTGCTGCCCTCAAGCCCGGC
TTGTGTGTTGGGTCGCCGTCCCCCTCTCCGGGGGGACGGGCCCGAAAGGCAGCGGC
GGCACCGCGTCCGATCCTCGAGCGTATGGGGCTTTGTCACATGCTCTGTAGGATTGGC
CGGCGCCTGCCGACGTTTTCCAACCATTCTTTCCAGGTTGACCTCGGATCAGGTAGGG
ATACCCGCTGAACTTAAGCATATATAAAACCCGGAGAGAACATTACCGAGTGCGGGTC
CGTTGGGCCAACCTCCATCCGTGTCTATTGTACCCTGTTGCTTCGGCGGGCCGCCGCT
TGTCGGCGCCGGGGGGGCGCCCTGCCCCGGGCCGGGCGCCAAACACCACGACAAAA
GATACAGACCGAGATGCTTGTATTAATGCTCCCCTACCGTCATTTACCTTTCAGAGGGG
TTTTGTGGGCTCCCGCCCCAAAAAAAAAAACCCCCAAATGGCCAATAACTAAATGTTG
AAATTTGCAAAATTTTTGTAAAATCCCAGTTCTTTTAAAAACCCTTTTCCCCCGCGGGT
TTTTTGGGGGGGGGGGGGCGCCCCACACACCCAAACGCTCCCCCCCGCCCGCGGTG
GGGGGGGTGGGGGACACCCCCCCCCCCCCCCGCGGCGCGGGGAAAAGGAGAAAAG
GAAGCCCCCCCCCCCCACCCCCACAGAAGGGGGGTGGTTGTGTTTTTTTTTTTTTTTT
TTATTAGCGCGGGCGGCCCACCTCCCCCCCCCCTCCCCCTCCCTCTCTTTCCTTTTTCTC
TTTGTGGGGAGGGAGCGCACACAATTAAAAAAAAAAAAAAAAAAAAAAAAGAAAAA
AAAAAAAAAAAAAAAAAAAAAAAAAAANAGNANTGAGGANGGCGAGAAGGAAAGA
NTTGGTTTTGTNTTGTTTGGTGTTATATATATCTCTTTAAATATCATTATAGTTTTATTNC
GAANTAAAGATACAGAGTAGACAATGAATAGTCACTNCTCCTCTTGACTGACTGAACG
AAATGTTTACCATAGTTAGTGAAGTAAAAAAGAAGAAGTTAATTGTGTTAACACCCGTC
GAATCGACGGCAGGCAGGTTGGTGAATG
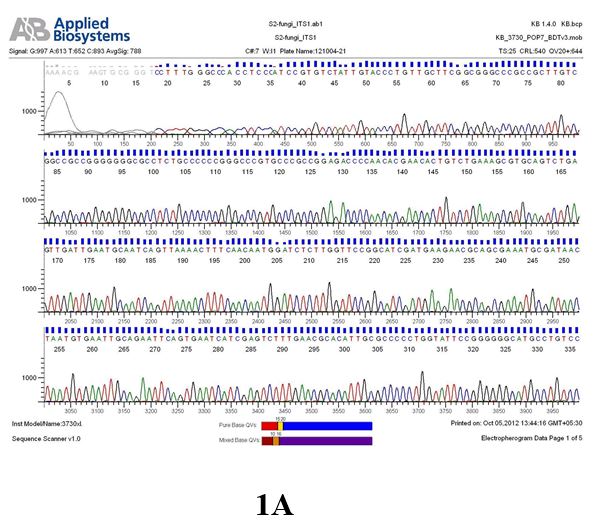
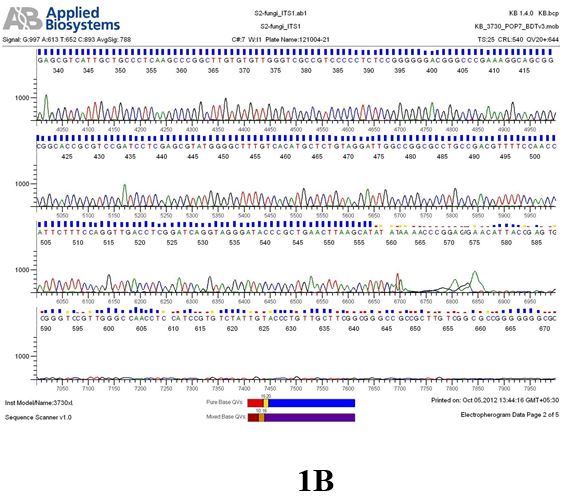
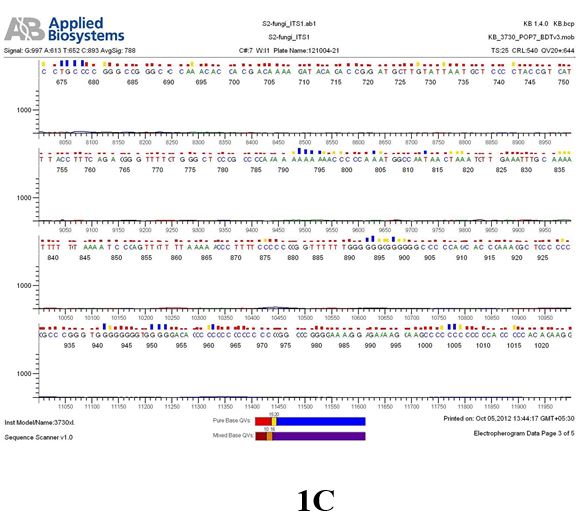
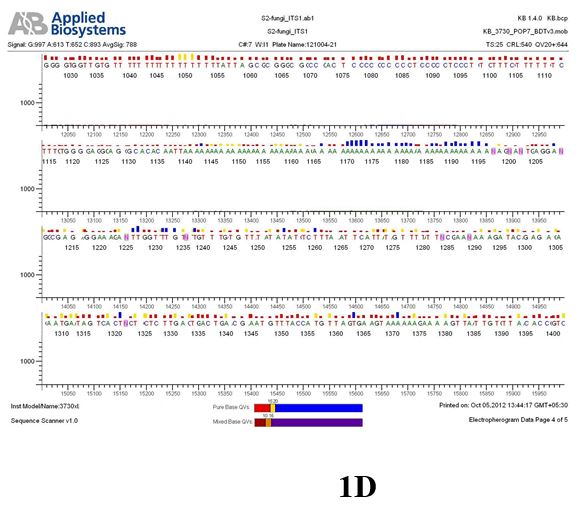
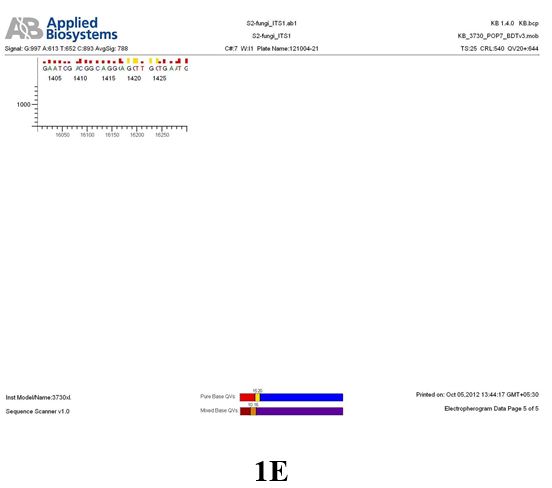
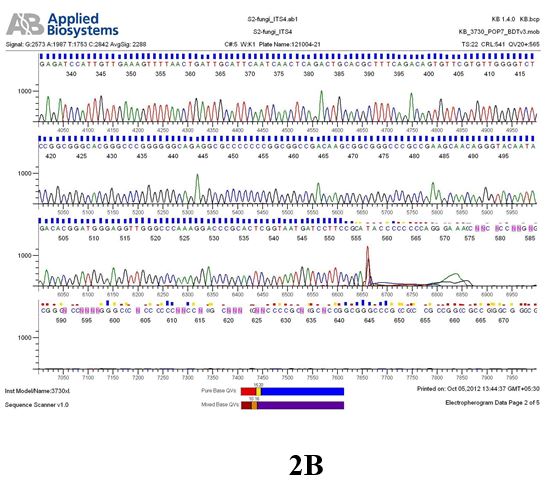
Result
The above sample of filamentous fungal culture DNA has been amplified in PCR. Hence, observed it’s sequencing pattern in forward, reverse and consensus sequences mention the pattern above. The quality of the sequencing results observed through chromatogram designed with two different internal transcribed spacer (ITS) polymerase chain reaction primer named as S2_fungi_ITS1.ab1 and S2_fungi_ITS4.ab1, both pairs with fungal ribosomal DNA which shows picks with four different colors position on accurate base pairs on chromatogram in forward and reverse direction. The sample S2 is identified to be Aspergillus candidus strain 085241 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence.
Discussions & Conclusions
Approximately 200 samples of frozen buffalo meat tested and 80% of the samples are found positive for both yeast and mould. It increases the risk associated with slaughter house daily practice of food processing and food hygiene. The fact outburst the survivability of psychrophilic organisms in extreme environment produces high risk in safe production of buffalo meat. Spores of fungus can withstand any kind of high tolerance of temperature, radiation and when found convenient and suitable condition spores germinate once again and lead to spoilage of meat.
As we know molds produces aflatoxins, ochratoxins, proteinase and lipase enzymes which on other hand contaminate the food and lead to heavy economical loss and spoilage. To reduce the effect of fungus and yeast activity irradiation of frozen meat packets are best option before releases to market. Few kind of yeast may used as probiotic agent or to use as bio-control to reduce the pathogenic organisms within flora of buffalo meat during processing, storing to consumption. Present study involves how air quality of slaughter house of different temperature is contaminated by yeast and mould which presented in this article by microbiological assessment149. Experiments of microbiological assessment check reveals that the tolerance of yeasts and moulds counts ranges from maximum 1.48 × 104cfu/gm to minimum 2.72 × 101cfu/gm. It is been seen as Asiatic Swamp Buffalo (Bubalus bubalis) a vital livestock which used from long time for food consumption which decreases the annual growth rate. Buffalo production need numerous management constraint including health issues biosecurity and transboundary diseases, nutritional dificits, low reproductive performance, high slaughter rate of pregnant buffalos, undeveloped trade and marketing system, limited verinary, potential climatic change and policy impact. During slaughter and processing may infect by fungal spores and leads to deterioration and spoilage. Hence, studies in this paper pinpointed for biosecurity of buffalo meat [20]. The partial gene sequence analysis of 18S rDNA sequencing test of two mold or fungus isolates from frozen buffalo meat sample reveals the identification of species and strains Such as. Aspergillus versicolor strain VA 18S ribosomal RNA gene & Aspergillus candidus strain 085241 18S ribosomal RNA gene.
Yeast & mold normally appear on meat in room temperature. The study displayed the high percentile of exported frozen buffalo meat were positive for Yeast & mold. The 18S rDNA sequencing analysis of mold also pinpointed and confirm the presence Aspergillus versicolor strain VA 18S ribosomal RNA gene & Aspergillus candidus strain 085241 18S ribosomal RNA gene. Such a huge concentration of contamination denotes the contamination label in slaughter house and during further processing of meat potential source of contamination. Therefore it is advisable to conclude to improve sanitary quality status of exported frozen meat in New Delhi, India slaughter house whether exported or locally produced [21-23].
Acknowledgement
This paper reviewed studies conducted by own financial support while working in a slaughter house in New Delhi, India. I am grateful to the Chancellor of Kalinga University and Principal & Ph.D. Supervisor of Shivaji Science College, Chickli, Maharastra for pursuing the work on Yeast and Mold. Even I am grateful
to the Management of Slaughter House in New Delhi for their co-operation towards completion of the work.
Bibliography

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!