Biography
Interests
Dr. Rahul Kamble, S.
Department of Laboratory Medicine, Hinduja Healthcare Surgical Hospital, Mumbai, India
*Correspondence to: Dr. Rahul Kamble, S., Department of Laboratory Medicine, Hinduja Healthcare Surgical Hospital, Mumbai, India.
Copyright © 2018 Dr. Rahul Kamble, S. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
The continuous variations in the bacterial resistance patterns in different geographical niches
necessitates updating of antimicrobial susceptibility profiles data regularly to guide clinicians in
choosing the appropriate empiric therapies. This retrospective study assessed the characteristics of
pathogens identified in clinical isolates from different clinical samples and their in vitro susceptibility
to commonly used antibiotics for multi-drug resistant (MDR) pathogens in our hospital settings.
A total of 212 samples from 210 patients treated for various bacterial infections during October
2016 to November 2017 were included in the study. Three antimicrobial agents had been used and
extended spectrum β-lactamases (ESBL) production was confirmed by double-disk synergy test.
Of the 212 isolates from 210 patients, 196 (92.0%) were Gram negative and 16 (8.0%) were
Gram positive organisms. Only 196 Gram negative isolates were included in the final analysis.
58.4% isolates were from urine, 12.3% from blood, 10.8% from pus and 18.5% from other sources.
Escherichia Coli was detected in half (50%) of clinical samples, followed by Klebsiella Pneumoniae
(25.8%) and Pseudomonas Aeruginosa (5.3%). The detection rate of the ESBL-producing isolates
was 20.7%. Resistance to piperacillin-tazobactam was most common among all the isolates. CSE-
1034(Ceftriaxone-sulbactam-EDTA-1034) had the greatest activity against both the ESBL and
non-ESBL E. coli (98.1%; 96.8%) and K. Pneumoniae (90.9%; 75%) respectively whereas both CSE-
1034 and meropenem exhibited similar level of activity against P. Aeruginosa (90.9%). All other
isolates except one isolate of enterobacter showed 100% sensitivity to all the three drugs.
This retrospective data suggest that CSE-1034 can be considered an important therapeutic option
for the treatment of both ESBL and Non-ESBL Gram-negative bacterial infections. A careful
monitoring of antimicrobial usage and resistance patterns should be done at regular intervals in
healthcare settings to combat the problem of antimicrobial resistance.
Introduction
The introduction of beta-lactam antibiotics heralded a new era in the fight against emerging bacterial resistance
mechanisms and comprised more than half of the antibiotics most widely prescribed for Gram-negative
infections [1]. However, the overuse and unrestricted consumption of these drugs for the long duration
across hospital and community settings has led to a surge in anti-microbial resistance, threatening the use of
the majority of large drug family [1]. Of particular concern are Asian countries that have witnessed a steady
rise in ESBLs producing Enterobacteriaceae, Metallo β-lactamases (MBLs) producing Enterobacteriaceae
and multidrug-resistant strains of P. Aeruginosa and A. Baumannii since last two decades [2]. The prevalence
of ESBL producers in India range from 28%-84% and MBLs range from 7-71% [3]. ESBLs are β-lactamase
enzymes capable of hydrolyzing various classes of antibiotics including penicillin’s, monobactams and other
several groups of β-lactam antibiotics, notably cephalosporins [4]. ESBL-producing organisms show crossresistance
to other groups of antibiotics also including fluoroquinolones, aminoglycosides, tetracyclines, and
trimethoprim/sulfamethoxazole [4]. The majority of ESBL producing organisms produce more than one
β-lactamase and strains producing multiple ESBLs are also reported.
Infections caused by multi drug-resistant Enterobacteriaceae are associated with increased morbidity and mortality compared to the ones caused by their susceptible counterparts [5,6]. This is mainly attributed to delayed active therapy supported by various studies demonstrating worse outcomes in patients with bacteremia due to Enterobacteriaceae with inactive empirical therapy [7,8].
The fact that antimicrobial resistance to first-line agents has been increasing makes the selection of empirical therapy more difficult [7,8]. Clinicians frequently face the dilemma between choosing one of those agents, which might not be active in some patients, or a drug with a very broad spectrum, knowing that this may further fuel the spread of resistance. For such decisions, various factors are considered in patients with infections potentially caused by Enterobacteriaceae, including individual risk factors, clinical severity and most importantly local epidemiology. The resistance patterns observed by a particular pathogenic species represent the sum of intrinsic and acquired mechanisms of resistance [9]. While the first are universally present, the latter ones are acquired and have heterogeneous prevalence and may be present only in some geographical areas. Thus, efficient reporting of the local epidemiology and the antibiotic susceptibility data of clinical isolates are helpful to clinicians for the appropriate management of patients. In the current study, the antibiotic susceptibility of Gram-negative clinical isolates, collected during the period of October 2016 to November 2017 against commonly used antibiotics was determined.
Materials and Methods
This study was carried on patients between 0 months to 89 years old, suffering from various bacterial
infections and treated in our hospital between October 2016 to November 2017. The clinical samples used
for pathogen isolation were urine, blood and pus. The sample collection and processing were done as per
standard microbiology laboratory operating procedures. Colony counts higher than or equal to 105 colony
forming units (CFU)/mL were considered significant.
Clinical isolates were identified based on Gram-staining, colony morphology, motility and different
biochemical reactions using standard techniques. The required clinical samples were collected in sufficient
amount in sterile containers aseptically. The collected specimens were inoculated or streaked on different
selective and non-selective culture media as per the standard microbiological procedures. Blood samples
collected in brain heart infusion (BHI) broth were incubated aerobically overnight at 37°C followed by subculturing
in the respective media.
In vitro susceptibility testing was done by Kirby-Bauer disk diffusion method for all the pathogen isolates as
per Clinical Laboratory Standard Institute (CLSI) guidelines. Discs of Ceftriaxone-sulbactam-EDTA-1034
(45μg), Meropenem (10μg), piperacillin/tazobactam (110μg) were purchased from Microexpress, a division
of Tulip Diagnostics private Limited, Goa, India. Inoculum suspension of 0.5 MacFarland standard turbidity
was prepared in a Mueller-Hinton broth (MHB, Hi-media, Mumbai, India) from a single colony picked
from 18-24 hours agar plates. A sterile cotton swab dipped into inoculum suspension and pressed against
the wall of tube above fluid level was streaked on Mueller-hinton agar (MHA) plate.
The swab was streaked two or more times at 60°C over the agar surface to ensure even distribution. After 3-5 minutes, antibiotic discs were implanted on to inoculated agar plates ensuring complete contact with agar surface. The discs were distributed evenly and a minimum distance of 24mm from center to center was ensured. Within 15 minutes, the plates were then inverted and incubated for 16-18 hours aerobically at 37˚C. Zones of inhibition were measured next day and were correlated with CLSI interpretive breakpoints to characterize them as sensitive, intermediate and resistant. CSE-1034, for which CLSI breakpoints are not available, interpretative breakpoints provided by the manufacturer were used. Criteria was >21mm- Sensitive, 14-20- Intermediate, ≤13- Resistance.
Results
A total of 212 culture positive clinical samples from 210 patients were included in this retrospective analysis.
The different clinical samples processed for pathogen isolation were urine, blood, pus, sputum, endo-tracheal
secretions and abdominal fluids. Of the total 212 clinical samples processed, the majority were urine (58.4%)
followed by blood (12.3%) and pus (10.8%). 59.5% isolates were from patients with UTI, 15% from wound
infections, 10.5% from patients with respiratory tract infections and 9.5% from patients with blood infections.
Further sub-group analysis on the basis of gender, had shown similar kind of pattern with urinary tract
infections (UTIs) predominant in both the genders accounting for 70% in females and 48.4% among males.
This was followed by wound infections (male; 22.1%: female; 10%) and respiratory tract infections (male;
12.6%: female; 8.2%). For other details, refer to Table no 1.
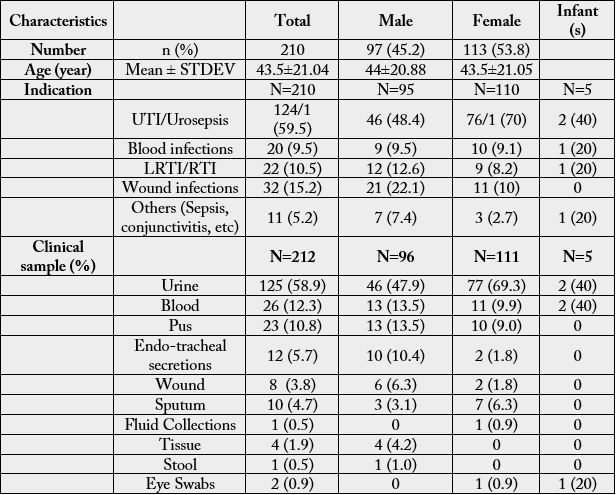
Among 212 bacterial isolates, 196 (92.0%) were Gram negative and 16 (8.0%) were Gram positive organisms.
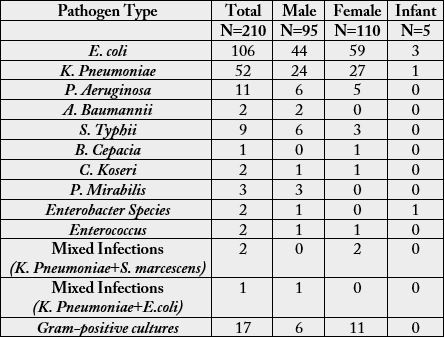
The detailed profile of various organisms isolated is shown in Table no 2. Only the gram-negative isolates were included in the final analysis. Around half of the isolates were of E. coli (50%) followed by K. Pneumoniae (25.8%), P. Aeruginosa (5.2%) and S. Typhii (4.2%). E. coli was predominant in both the genders followed by K. Pneumoniae (Table 2).
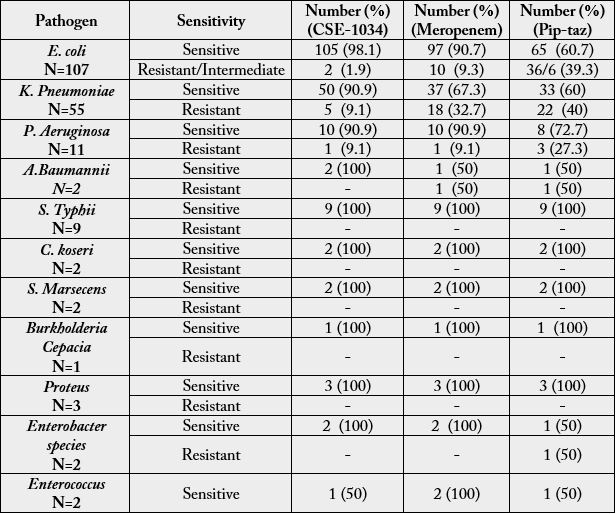
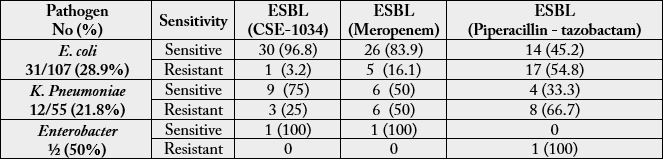
Antibiogram profile for all the pathogen isolates is shown in Table 3 and ESBL isolates in Table 4. Of the
107 E. coli isolates, 98.1% were susceptible to CSE-1034, 90.7% to meropenem and 60.7% to piperacillintazobactam.
31 (28.9%) E. coli isolates were ESBL producers. The sensitivity pattern observed for ESBL
producers was although in line with all pathogen isolates but the number varied significantly in different
antibiotic groups. 96.8% of ESBL E. coli isolates were sensitive to CSE-1034 compared to 83.9% to
meropenem and significantly low (45.2%) to piperacillin-tazobactam.
The percentage of isolates positive for different pathogens by infection type, and the proportion of ESBL producing isolates are presented in Table 5.
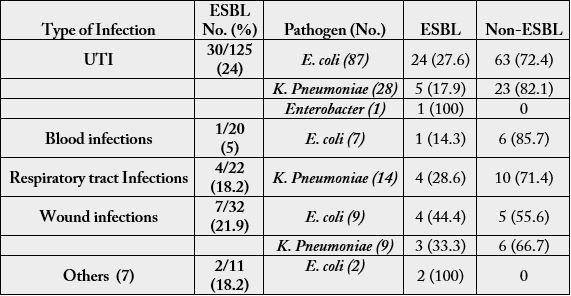
Overall, 55 (25.8%) isolates were identified as K. Pneumoniae and 11 (5.1%) as P. Aeruginosa. The meropenem
susceptibility rate of K. Pneumoniae was 67.2% (37/55) and P. Aeruginosa was 90.9% (10/11). The least
sensitivity among all the isolates was shown towards piperacillin-tazobactam; K. Pneumoniae (60%; 33/55)
and P. Aeruginosa (72.7%; 8/11).
CSE-1034 had a better activity than meropenem and piperacillin-tazobactam against both the pathogen isolates; 90.9% towards both K. Pneumoniae (50/55) and P. Aeruginosa (10/11). 12/55 (21.8%) of K. Pneumoniae isolates were ESBL producers whereas none of the P. Aeruginosa isolates was ESBL producer.
The sensitive percentage of ESBL producing K. Pneumoniae to CSE-1034 was 75% which is significantly high compared to meropenem (50%) and piperacillin-tazobactam (33.3%).
Overall, 6% isolates of S. Typhii, 2.9% of A. Baumannii and C. Koseri, 1.4% of Proteus, 0.9% each of S.
Marsecens and Enterococcus were isolated. All these isolates showed 100% sensitivity to piperacillin-tazobactam,
meropenem and CSE-1034 and none was reported as ESBL. Out of two isolates of Enterobacter, one isolate
reported as ESBL producer was resistant to piperacillin-tazobactam.
Discussion
Although, various guidelines are available for the management of bacterial infections, however adoption
of these guidelines at the hospital level is challenged by difference in the predominant pathogens, their
sensitivity pattern and health care-associated economic conditions that varies with different geographical
locales [7,10]. Therefore, the audit and analysis of the microbiological isolates and their sensitivity pattern
is indispensable to clinicians for right empiric therapy. In this study, it was found that UTI was the most
prevalent infection in both the genders accounting for almost half of isolates. Based on gram staining, Gramnegative
pathogens were predominately isolated (92%) in all clinical samples analyzed with urine being
the most processed one followed by blood and pus. The results obtained in this study are in concordance
with other studies conducted in other parts of India and Asian sub-continent. A recent retrospective antimicrobial
susceptibility study conducted at a tertiary care center in Delhi, India has reported 76.4% isolates
as gram negative [11]. Similarly, Karanwal et al. [12] have reported same pattern from Ahmedabad with
Gram-negative isolates comprising 78% of the total isolates and Prabhash et al. [13] from Mumbai (Gramnegative
organisms comprising 68.1% of blood cultures). Bacteriological profile in India differs from that in
the west. Although, the gram-positive pathogens predominate in Europe, on the contrary more incidences
of gram-negative pathogens are reported in Asia. Among the identified Gram-negative isolates, E.coli was
the most frequent pathogen in both the genders accounting for 42% of total isolates. Other pathogens
identified were K. Pneumoniae (27.3%) and P. Aeruginosa (9.3%). Although E coli was the leading cause of
UTI in both the sexes, its proportion was significantly higher in women compared to men. In accordance
to our results, various studies have reported Gram-negative bacilli as the most common pathogens causing
UTIs in both men and women with a ratio of 1:2, and E. coli being the most prevalent type [14,15].
An important mechanism of antibiotic resistance in gram negative pathogens is through ESBL production. ESBLs are plasmid mediated and derived from class A b-lactamases, and their spectrum includes oxyimino-β- lactams including penicillin’s, aztreonam, monobactams, cephalosporins (with the exception of cephamycin’s) [4]. In addition, ESBL-producing organisms frequently show cross-resistance to other classes of non-β- lactam antibiotics including trimethoprim-sulfamethoxazole, aminoglycosides, and fluoroquinolones, thus making the treatment of these infections a therapeutic challenge. ESBLs are commonly found in Gramnegative bacterial isolates mainly in Enterobacteriaceae and have been reported worldwide [16-18]. Consistent with these results, all the ESBL-producing pathogens were from Enterobacteriaceae family and none was reported from non-Enterobacteriaceae family. Further, in Enterobacteriaceae family, E.coli and K. Pneumoniae leaped out as the significant ESBL producers and contributed 28.9% and 21.8%, respectively. In India, Sentry surveillance study has reported the prevalence of ESBLs from 62% to 100% in E. coli and Klebsiella spp. isolated from respiratory infections, blood stream infections, skin and soft tissue infections [11], The proportion of ESBL isolates also varied with the infection type: the prevalence of EBSL-producing isolates was highest from UTI cases. The ESBL isolates from wound infections was 15.9% followed by respiratory infections (9.1%).
Susceptibility results showed that the resistance rate of ESBL-producing isolates against used antimicrobial drugs was higher than the rate observed in ESBL-negative isolates. A high rate of resistance of both ESBL and non-ESBL isolates was observed to piperacillin-tazobactam. The indiscriminate consumption of piperacillin-tazobactam could be one of the vital reasons for the high antimicrobial resistance reported towards the normally recommended second line of treatment in our hospital. Singh et al. have reported 22% of E. coli isolates, 50% of Klebsiella and 35% of P. Aeruginosa resistant to piperacillin-tazobactam in their study.
A 98.1% and 96.8% sensitivity of Ceftriaxone-sulbactam-EDTA-1034 (CSE-1034) was reported against ESBL-negative and ESBL-producing E. coli and 90.9% and 75% against K. Pneumoniae respectively in our study. The anti-bacterial effect of this compound on non-ESBL and ESBL-producing isolates was superior to meropenem; 90.7 & 83.9% for E. coli and 67.3% & 50% for K. Pneumoniae, respectively. Our study has shown that non-ESBL P. Aeruginosa showed 90.9% sensitivity rate. The emergence of carbapenem-resistant strains, which ranges from 9-32% in this study is a matter of big concern as carbapenems are considered as the last resort drugs for ESBL strains. A significantly higher incidence of carbapenem-resistant Gramnegative bacteria has also been reported by Ghosh et al. [19] from AIIMS, Delhi. Similarly, Singh et al. [11] have reported that 15-22% of the gram-negative isolates were metallo-beta lactamases (MBLs)in their study. Interestingly, a significant number of both ESBL and non-ESBL isolates were sensitive to CSE-1034. The high sensitivity of gram-negative pathogens to CSE-1034 has been reported by several other studies also [20] [21]. 100%, 64% and 63% of ESBL producing A. Baumannii, K. Pneumoniae and E. coli were reported susceptible to CSE-1034 in an antimicrobial susceptibility pattern study conducted by Sahu et al. [22]. In the same study, 89%, 60%, 42% and 41% of MBL producing isolates of A. Baumannii, E. coli, P. Aeruginosa and K. Pneumoniae, respectively were susceptible to CSE-1034.
Conclusion
Our analysis suggests that pathogen isolates from various clinical sources demonstrated a varied rate of resistance to different antimicrobials in our hospital. CSE-1034 and carbapenems remained the most effective drugs against gram-negative pathogens. This finding assumes significance in the backdrop of formulating empirical therapy for patients suspected of suffering from multi drug resistance (MDR) infections. As CSE-1034, a beta-lactam/beta-lactamase combination was equally effective against various infections, it can be a drug of choice for MDR bacterial infections to combat the overuse of carbapenems and reduce the selection
pressure on carbapenem-resistant strains. Moreover, a high resistance pattern observed in this study warrants
a careful monitoring of anti-microbial use in hospitals.
Bibliography

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!