Biography
Interests
Olorunjuwon Omolaja Bello1*, Mathew Olujenyo Oni2, Temitope Kudirat Bello3 & Bamidele Precious Awopetu1
1Department of Biological Sciences, University of Medical Sciences, Ondo City, Nigeria
2Department of Biological Sciences, Adeleke University, Ede, Osun State, Nigeria
3Department of Biological Sciences, Elizade University, Ilara-Mokin, Ondo State, Nigeria
*Correspondence to: Dr. Olorunjuwon Omolaja Bello, Department of Biological Sciences, University of Medical Sciences, Ondo City, Nigeria.
Copyright © 2021 Dr. Olorunjuwon Omolaja Bello, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
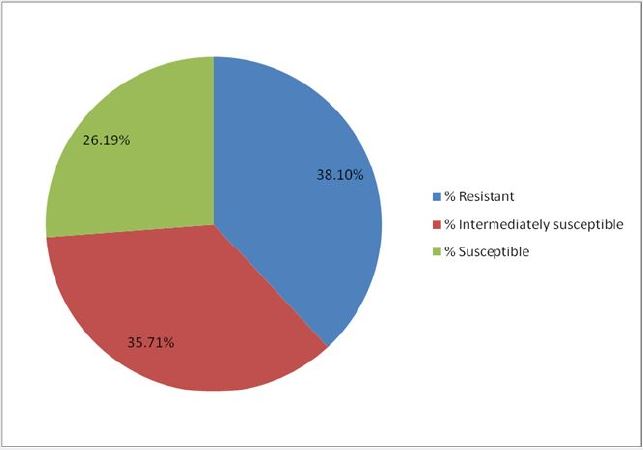
Antibiotics used in the treatment of dental caries infection are often wrongly prescribed which resulted in the increased resistance in bacteria associated with teeth infections. The purpose of this research was to screen infected teeth of dental patients attending University of Medical Sciences Teaching Hospital (UNIMEDTH), Ondo City, for associated antibiotic-resistance bacteria. Swab samples were taken from consented dental patients at the UNIMEDTH and analyzed microbiologically using standard procedures. Nutrient agar and blood agar media were used to cultivate the total viable bacteria and fastidious organisms, respectively. The most probable identity of bacterial isolates was determined by the conventional biochemical tests and confirmed with API 20 E and API 20 NE. Antibiotic sensitivity of bacterial isolates was investigated using the agar disc diffusion method in accordance to standard procedures. The bacterial isolates were obtained and identified as Staphylococcus epidermidis 4(28.57%), Staphylococcus sp 3(21.42%), Streptococcus sp 3(21.42%), Enterococcus sp 3(21.42%) and Pseudomonas sp 1(7.14%). It was found that 7 (50%) of the bacterial isolates exhibited resistance to amoxil, 6(42.86%) showed resistance to streptomycin, nalidixic acid, chloramphenicol and rifampicin. However, 5(35.71%) to tarivid, ciprofloxacin, erythromycin, ampicillin and norfloxacin while 4(28.57%) displayed resistance to gentamicin and levofloxacin. Exactly 38.1% of the bacterial isolates exhibited resistance, 35.71% were intermediately susceptible while 26.19% were susceptible to the investigated antibiotics. This study revealed that the bacteria isolated from the infected teeth were resistant to most conventional antibiotics investigated. Further studies are required to determine their susceptibility to extracts of selected medicinal plants.
Introduction
The hardest substances constituting any part of the human body are the teeth. Every tooth’s crown projects into the mouth with its root descending down the gum line and into the jaw [1]. Apart from being essential for chewing, they play an important role in speech. Part of the teeth includes the enamel, (the hardest, the white outer part of the tooth, it is mostly made of calcium phosphate and a rock hard mineral), the dentin (a layer underlying the enamel, when the enamel is destroyed, heat or cold can enter the tooth through these paths and cause sensitivity or pain), pulp, cementum, periodontal ligament. A normal adult mouth has 32 teeth, which includes the (incisors=8, canines=4, premolars=8, molars=4, wisdom teeth=4) [2].
The oral cavity has a lot of features that make it a distinct microbial habitat. The human oral cavity harbors a complex microbial community of a diverse range of microbes and it comprises several species of bacteria, fungi, and protozoa. The oral cavity is normally relatively stable but under certain circumstances can cause major oral disease and they are considered to be one of the major public health challenges as related to their high rate of occurrence around the globe. These diseases include dental caries, periodontitis, gingivitis, tooth loss, etc. The destruction of the calcified tissue of the teeth is known as dental caries and it happens as a result of the presence of different bacteria in the oral cavity that can use the fermentable carbohydrates that surrounds the tooth for a long period of time. It has been estimated that about 700 species of bacteria are capable of inhabiting the oral cavity [3].
Dental caries is one of the major and most prevalent chronic diseases affecting children and adults globally. People are prone to developing this disease at a point in their lifetime. The disease develops in both the crowns and roots of teeth, and it can arise in early childhood as an aggressive tooth decay that affects the primary tooth of infants and toddlers. The World Health Organization (WHO) [4] estimated that oral diseases affect about 3.5 billion people globally and that over 530 million children suffered infection of the primary teeth. Dental caries happen when plaque forms on the surface of a tooth and converts free sugars contained in food and drinks into acids that destroy the tooth over time. Continuation of high intake of free sugars, inadequate exposure to fluoride, and inadequate removal of plaque by tooth brushing can lead to caries, pain, infection, and sometimes tooth loss.
The microbial community of dental caries is diverse and contains a number of facultative and obligateanaerobic bacteria. Dental caries affect humans in several ways ranging from the presence of tooth pain, discomfort to infection, and/or dysfunction of the stomatognathic system, depending on its severity [2]. These could in turn limit the ingestion of nutritional foods affecting growth, especially in children, learning, communication skills, and recreational activities. This is one of the major health burdens for several countries. Personal hygiene and dietary modification had been recommended as means of combating this menace [5].
The oral cavity of human is a complex ecosystem which comprises both acid-producing and acid-tolerant bacteria. Almost 700 different bacterial species are associated with human oral cavity, and about 200-300 species have been linked with dental plaque. The interaction between bacteria and its surrounding epithelium is an acute element in bacterial infections. Dental plaque is the sticky substance that adheres to the surface of the teeth. It is a complex biofilm that causes dental caries. Dextran, produced by the dietary sucrose fermentation by Streptococcus mutans, is responsible for the stickiness of the plaque [6].
S. mutans initiate enamel decalcification as a result of the production of lactic acid during fermentation process. This plays a role in the initiation of enamel caries. The occurrence of dental plaque is usually as a result of the interaction between the plaque attachment to the surface of the tooth and the physical shear forces that dislodge and remove the plaque. If the dental plaque is not removed properly, tooth decay will flourish [7]. The mature dental plaque is embedded in a matrix of bacteria and host polymer that includes proteins, DNA secreted by cells, and polysaccharides [8].
The main factors associated with dental caries include bacteria, susceptible tooth surface, time, and fermentable carbohydrates. However, certain behavioral and sociodemographic factors also increase the risk of caries and these include improper tooth brushing habits, poor oral hygiene, age, plaque, and sugar-containing drinks [8]. These diseases can be treated by brushing teeth, flossing teeth, rinsing teeth, teeth cleaning, tooth filling, root canal, root extraction, braces, mouth guard, dental sealants, and teeth whitening [2].
The use of antibiotics has changed the natural evolution of bacteria by reducing susceptible pathogenic populations and increasing resistant populations. Resistance is most times linked with a fitness cost as a result of the genetic burden needed for resistance and which could be deleterious in antibiotic-free environments. In this case, restriction of the use of antibiotics has been proposed, with the aim of eradicating resistant bacteria. However, the genetic background of resistant pathogens allows them to persist in the presence of minimal concentrations of antibiotics or even in the absence of these [9]. This study, therefore, aimed to screen infected teeth of dental patients attending the University of Medical Sciences Teaching Hospital, Ondo City for associated antibiotic-resistance bacteria.
Materials and Methods
The project study area was the University of Medical Sciences Teaching Hospital, Ondo City, Ondo State,
Nigeria.
Permission to collect swab samples for the research was sought from the Chief Medical Director, Dental
Unit, University of Medical Sciences Teaching Hospital, Ondo City, Ondo State, Nigeria. The patients
were informed of the purpose of the research and all information collected during the study was considered
confidential.
Specimens were collected from the dental units at the University of Medical Sciences Teaching Complex
in Ondo City. Samples were collected by swabbing the teeth of infected patients using a sterile swab sticks.
The swab sticks were rubbed over the surface of the infected teeth, and which were immediately transferred
to the Biological Sciences laboratory at the University of Medical Sciences, Ondo.
Samples from the infected teeth of the patients were inoculated on nutrient agar media using the streak
method and the plates were incubated at 37ºC for 18-24 hours, and the total viable bacteria were counted. The
isolates were subcultured aseptically on blood agar media to encourage the growth of fastidious organisms.
The blood agar plates were also incubated at 37ºC for 18-24 hours.
The isolated bacteria were identified by Gram staining and the conventional biochemical tests which
included catalase, coagulase, citrate utilization, methyl red, urease, indole and Voges Prokauer tests. Motility
and sugar fermentation tests were also carried out while further identification of the isolates was carried
out using API 20E and API 20NE to confirm the families Enterbacteriaceae and non-Enterobacteriaceae
respectively [10].
Mueller Hilton agar was prepared and it was sterilized in an autoclave at 121ºC for 15 minutes, it was
poured into Petri dishes and allowed to cool. Bacteria were streaked on the surface of the agar plates and
disks of filter paper with standard concentration of antibiotics were applied on the surface of the plates.
The antibiotics investigated in this study were tarivid, ciproflox, gentamycin, streptomycin, nalidixic acid,
ampicillin, norfloxacin, erythromycin, chloramphenicol, levofloxacin and rifampicin. As the bacteria grew
on the surface of the plates, the antibiotics diffused from the paper disc out into the agar plates. The plates
were incubated at 37ºC for 24 hours. A clear area of no growth around the particular disc meant that the
isolate was susceptible to a particular antibiotic. The zone around an antibiotic disc that had no growth was
referred to as the zone of inhibition since this approximates the minimum antibiotic concentration sufficient
to prevent the growth of the test isolate. The zone was then measured in mm and compared to a standard
interpretation chart to categorize the isolates as susceptible, intermediately susceptible or resistant [11].
Results
Table 1 shows the demographic data of dental patients that participated in the study. Ten dental patients
consented to the study out which 6 (60%) were female and 4 (40%) were male. The age range was 15 to 28
years while the ranges of the weight and height were 42 to 59kg, and 5.1 to 5.7 ft, respectively.
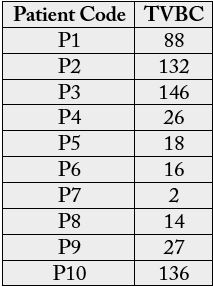
The total viable bacterial count (TVBC) of direct swab culture from infected teeth of dental patients investigated is shown in Table 2. The TVBC ranged from 2 to 146 colonies per plate. Table 3 shows the morphological and biochemical characteristics of bacteria isolated from infected teeth of dental patients. A total of fourteen isolates were obtained and characterized into 4 genera but 5 species of microorganisms. The most probable organisms identified were Staphylococcus aureus, Streptococcus mutans, Staphylococcus epidermidis, Pseudomonas aeruginosa and Enterocococcus faecalis. The percentage frequency of the bacterial isolates is given Figure 1. S. epidermidis showed the highest percentage occurrence of 28.57%, and followed by S. aureus (21.42%), S. mutans (21.42%), E. faecalis (21.42%) while the lowest was P. aeruginosa (7.14%).
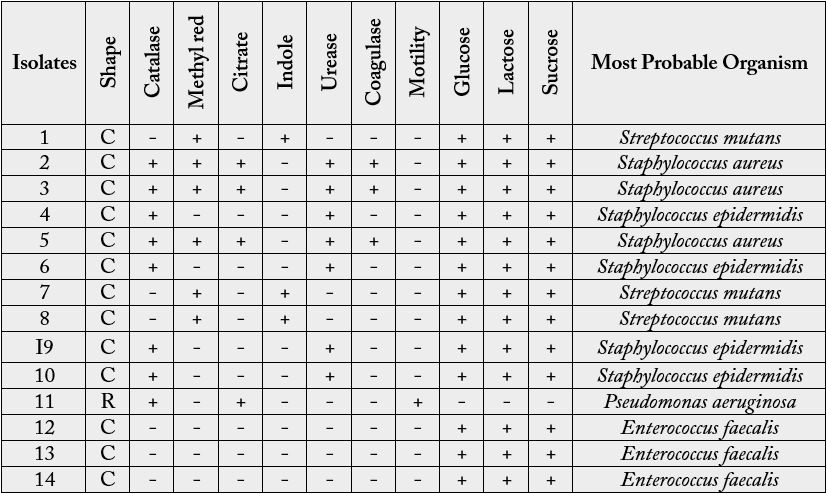
Key: - Negative; + Positive; C- Cocci; R- Rod
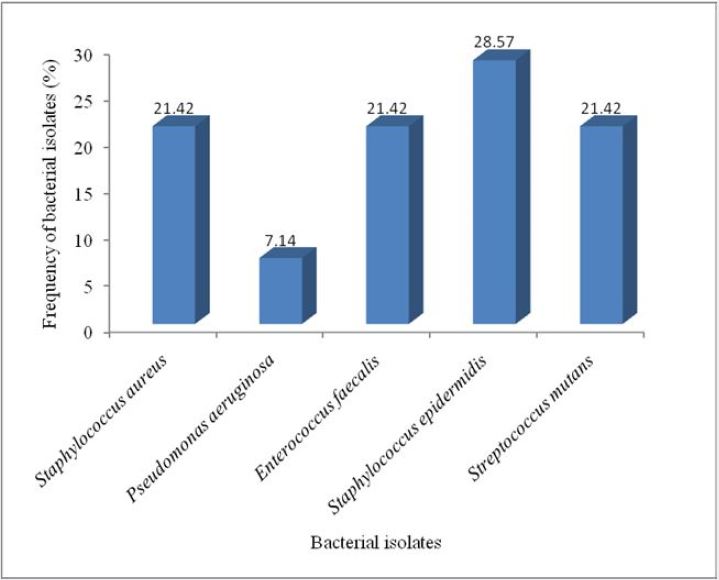
Table 4 shows the antibiotic sensitivity profile of the isolated bacterial from infected teeth of dental patients. The antibiotics investigated exhibited different spectra of activities against the bacterial isolates. The zones of inhibition were interpreted as susceptible (S), intermediately susceptible (I) and resistant (R) in line with the CLSI [11]. The percentage resistance profile of bacterial isolates from infected teeth of dental patients is shown in Figure 2. Exactly 38.1% of the bacterial isolates exhibited resistance, 35.71% were intermediately susceptible while 26.19% were susceptible to the investigated antibiotics.
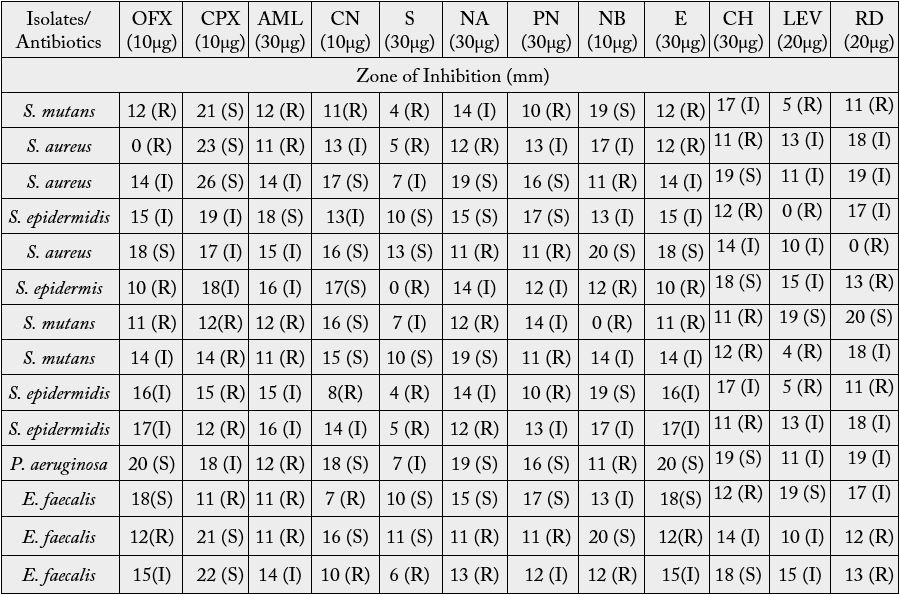
Key: S- Susceptible; I- Intermediate; R- Resistance; OFX- Tarivid; CPX- Ciproflox; CN- Gentamycin; S- Streptomycin; NA- Nalidixic Acid; PN- Ampicillin; NB- Norfloxacin; E- Erythromycin; CHChloramphenicol; LEV- Levofloxacin; RD- Rifampicin
Discussion
S. epidermidis, S. aureus, S. mutans, E. faecalis and P. aeruginosa were encountered in the present study. This
study encountered some of the bacteria reported by Bello et al. [12] who reported Streptococcus mutans,
Klebsiella pneumoniae, Staphylococcus albus, Proteus vulgaris and Pseudomonas aeruginosa. However, K.
pneumoniae, Staphylococcus albus and Proteus vulgaris were not encountered in this study.
S. epidermidis was the most frequent bacterium associated with the infected teeth investigated in this study with a percentage frequency of 28.57%. This finding differs from the study of Bello et al. [12] who reported S. mutans as the most frequent bacterium. This result was buttressed by findings of Olajokun et al. [13] who isolated S. mutans (45.6%), Lactobacillus spp (41.2%) and S. aureus (13.2%) from carious lesion. However, S. epidermidis, E. faecalis and P. aeruginosa were not encountered in their study. A study carried out at General Hospital Minna, Nigeria reported S, aureus as the most prevalent organism, followed by S. mutans and then Lactobacillus spp. which is contrary to the findings in the study where S. epidermidis was the most prevalent (28.57%), followed by S. aureus (21.42%), S. mutans (21.42%), E. faecalis (21.42%) and then P. aeruginosa (7.14%).
S. mutans is considered the main organism responsible for human dental caries. Certain factors like the tolerance of frequent and rapid environmental fluctuations and its frequent tolerance, the ability to form biofilms, and metabolizing carbohydrates are contributing factors to the virulence of this bacterium. S. mutans is also associated with bacterial endocarditis. The inflammation of heart valves and bacterial endocarditis had been linked with S. mutans. Synthesis of the extracellular polysaccharides by S. mutans from sucrose through glucosyltransferases (GTFs) is considered another important virulence factor that causes caries in humans [8].
This not only facilitates the adhesion and accumulation of the organism on the tooth surface but also provides protection against host immune defenses along with the provision of increased resistance against antibiotics and gene expression. This combination of virulence properties allows the S. mutans to colonize the surface of the tooth and modify the nonpathogenic to the highly cariogenic dental biofilm that ultimately leads to caries formation [14].
The impact of several different classes of medications and the effects that those medications had on oral health had been described [15]. A number of methods are used in oral hygiene to prevent and cure oral diseases. In many African homes, teeth are cleaned in the morning by chewing the root or slim stem of certain plants until they acquire brush-like ends [16]. Bello et al. [5] in their study on used toothbrushes mentioned that sharing of toothbrushes could result in an exchange of body fluids and/or microorganisms between the users of the toothbrush, placing the individuals involved at an increased risk for dental infections. They mentioned that the practice should be of particular concern for persons with compromised immune systems or existing infectious diseases. It was found that 7 (50%) of the fourteen bacterial isolates exhibited resistance to amoxil, 6 (42.86%) showed resistance to streptomycin, nalidixic acid, chloramphenicol, and rifampicin. 5 (35.71%) to tarivid, ciprofloxacin, erythromycin, ampicillin, and norfloxacin while 4 (28.57%) displayed resistance to gentamicin and levofloxacin.
The increasing prevalence and spread of antimicrobial-resistant strains of bacteria is evidently threatening the capacity of treating infectious diseases. In effect, this poses a significant burden on public health. The prevalence and/or the occurrence of multidrug-resistant/antimicrobial resistance microorganisms in clinical environments such as hospitals have been earlier reported [17]. Antibiotics resistance is not uniform with all strains, but even a low prevalence can change rapidly through a combination of the action of transferable resistance determinants and frequent exposure to antibiotics.
Erythromycin exerted antimicrobial potency against many of the bacterial isolates in this study which was buttressed by the reports of Bello and Bello [10] and Hardman et al. [18] where over half of the organisms isolated were sensitive to the same class of antibiotics. Contrary to the report of Yadav [19] that strains of S. mutans and S. aureus were totally resistant to erythromycin, 3 of 14 (2.14%) isolates were susceptible while 6 (42.9%) showed intermediate susceptibility (Table 4). Erythromycin is a macrolide-based antibiotic that reversibly binds to the 50s ribosomal subunit to inhibit the synthesis of protein [20]. Some strains of S. mutans showed susceptibility and intermediate susceptibility to gentamicin in this study, contrary to the report of Istifanus [21] who reported that all strains of S. mutans investigated in their study were resistant. Kariminik [22], however, reported that all S mutans were sensitive to gentamycin. Gentamicin belongs to the aminoglycoside class of antibiotics. The high potency exerted by the gentamicin against bacterial isolates could be associated with the mechanism of action of this class of antibiotics which enables it to bind irreversibly to the 16S rRNA subunit of the 30S ribosome, resulting in inhibition of bacterial protein synthesis. This finding is supported by the reports of Barber et al. [23], Mhondoro et al. [24], and Breijyeh et al. [25].
A high percentage of the bacterial isolates (42.9%) were resistant to chloramphenicol. Yadav [19] also reported that only a small percentage of the isolates encountered in his study were sensitive to chloramphenicol. Chloramphenicol belongs to the phenicol class whose mode of action is to interfere with bacterial protein synthesis. The production of chloramphenicol acetyltransferase (CAT) is responsible for the resistance of bacteria to chloramphenicol while some resistance occurs as a result of the inability of certain bacteria to reach their target sites. Although dental undergraduates are taught that most oral infections may be treated by surgical or mechanical means, without the use of antibiotics, dentists are prescribing millions of pounds’ worth of antibiotics every year. The therapy used is broad-spectrum antibiotics [26].
However, cultivating the microorganisms associated with dental infection and carrying out susceptibility testing to aid diagnosis and rational choice of antibiotic does not normally precede prescription. This implies that broad-spectrum antibiotics are often prescribed for a range of infections associated with tooth decay which they may not be required. This is, therefore, one of the major causes of the development and spread of antibiotic-resistant pathogens associated with dental infections, and which poses public health risk in relation to increased cost of care, morbidity, and future treatment choice.
Conclusion
This study revealed that the bacteria isolated from the infected teeth were resistant to most conventional
antibiotics investigated. The challenge and public health threat of antibiotic resistance in oral flora requires
further investigation with larger subjects. A systematic assessment of drug-prescribing practice should be helpful, while it should be borne in mind that prescriptions are neither a direct measure of the rate of
antibiotic consumption nor a measure of the rate of emergence of resistance, which is multifactorial.
Conflicts of Interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Bibliography

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!