Biography
Interests
Eswari Beeram
Department of Biochemistry, Sri Venkateswara University, Tirupati, Andhra Pradesh, India
*Correspondence to: Dr. Eswari Beeram, Department of Biochemistry, Sri Venkateswara University, Tirupati, Andhra Pradesh, India.
Copyright © 2019 Dr. Eswari Beeram. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
MALDI or Matrix Assisted Laser Desorption /Ionisation is well known technique which is generally used for analysis of proteins. Now it is well known for carbohydrates and lipids and as well as for chemical compounds. Linear mode is generally used for compounds which are easily fragmented where as reflection mode results in better resolution as it doubles the path of ions before reaching the detector. Positive mode is preferred for positive ions and negative mode for negative ions. Phosphoglycerolipids like phosphatidyl serine and phosphatidyl ethanolamine possess negative charge at physiological pH so its identification is preferable at negative mode. RNase A is a basic protein and has positive charge at physiological pH as it possesses pI of 9.6. Above the neutral pH aminoacids possess negative charge and below the neutral pH aminoacids possess positive charge. When RNaseA is subjected to MALDI analysis peptide of 14-17 aminoacids predominates in both positive and as well as in negative linear mode.
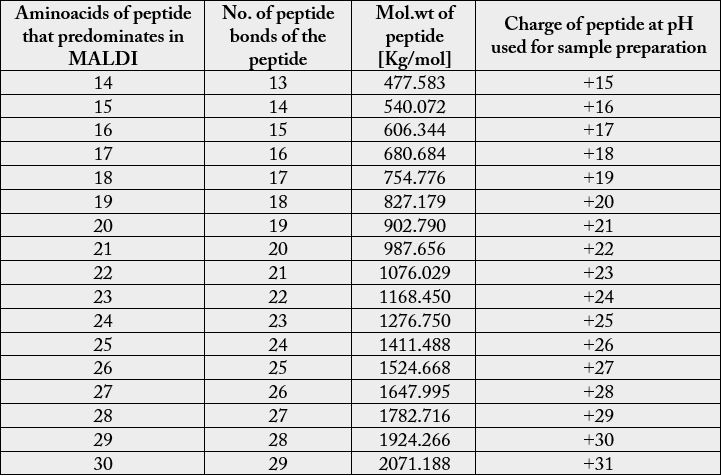
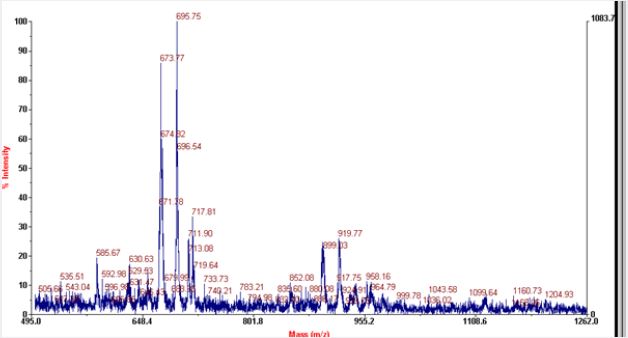
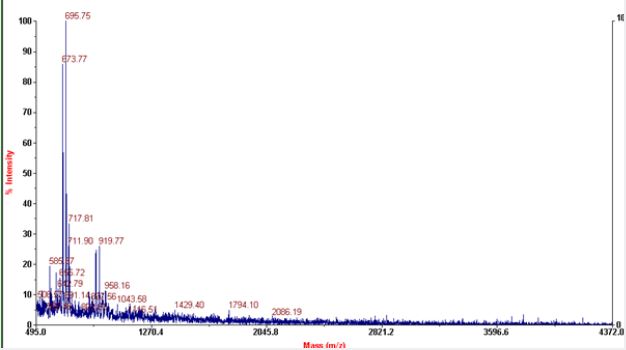
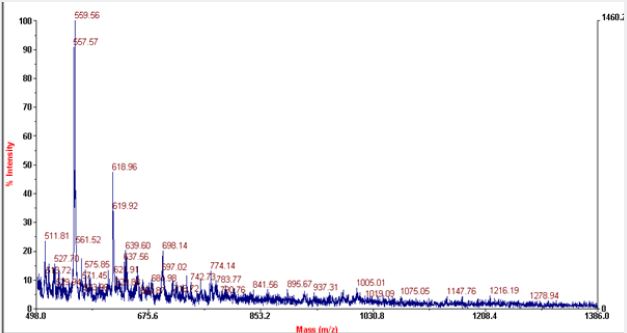
Most of the commonly used matrices in MALDI include sinapinic acid, CHCA and DHB. Sinapinic acid is mostly used for peptides having mol.wt above 10kda. Mol.wt of RNaseA is found to be 13.7kda with average mol.wt. of aminoacids it contain, but the actual mol.wt. of the protein is found to be 211,172Kg/ mole. It consists of 124 aminoacids and with peptide bonds of 123. It consists of four disulfide bond residues at 26-84, 40-95, 58-110, 65-72. The secondary structure of RNase A consists of 36 hydrogen bonds involving main chain atoms. The most prominent peaks in MALDI includes peptides of 14-17 aminoacids. RNase A possess ‘+’ charge at the pH of solvents used as the final pH falls around 3 after addition of TFA to acetonitrile. As there is prominent peaks in negative linear mode, so may be the protein loose some of the protons and convert to negative ions during fragmentation. Peptides of 15 -24 aminoacids with difference of 1 aminoacid predominates in both positive and negative linear modes. In negative linear mode peptides having mol.wt. above 2071.188kg/mole are absent where as in positive mode peptides that give peaks mol. wt. above 1278 are absent.

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!